Construction and validation of an anoikis-related long non-coding RNA-based prognostic model for head and neck squamous cell carcinoma
- PMID: 40792140
- PMCID: PMC12335720
- DOI: 10.21037/tcr-2024-2520
Construction and validation of an anoikis-related long non-coding RNA-based prognostic model for head and neck squamous cell carcinoma
Abstract
Background: As a unique form of apoptosis, anoikis significantly influences tumor biology. Studies have revealed the diverse roles of long non-coding RNAs (lncRNAs) in cancer signaling pathways; however, the prognostic significance of anoikis-related long non-coding RNAs (ARLncs) in head and neck squamous cell carcinoma (HNSCC) remains unexplored. Therefore, this research was undertaken to establish a risk model and assess its predictive ability for prognosis and immune landscape in individuals with HNSCC.
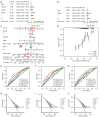
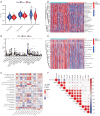
Methods: Data on HNSCC were retrieved from The Cancer Genome Atlas (TCGA). Anoikis-associated genes were acquired from GeneCards, followed by identification of ARLxncs using Pearson correlation analysis. A total of 268 ARLncs from HNSCC samples were extracted from TCGA, and highly relevant ARLncs were identified using Pearson analysis. These ARLncs were subjected to comprehensive bioinformatics analyses, including univariate Cox regression and least absolute shrinkage and selection operator analyses, and an overall survival (OS)-score and OS-signature were generated.
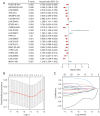
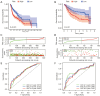
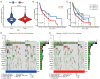
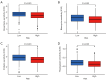
Results: Based on the risk score, patients with HNSCC were stratified into high- and low-risk subgroups to assess the differences in pathway enrichment, prognosis, immune infiltration level, tumor mutation burden, and drug susceptibility. TCGA-HNSCC samples were divided into two subtypes (clusters 1 and 2), with patients in cluster 2 exhibiting worse prognosis and higher levels of tumor-infiltrating lymphocytes (TILs) than patients in cluster 1. Subsequently, we constructed a valid prognostic risk model comprising 12 ARLncs in HNSCC that demonstrated efficacy in predicting prognosis. Patients with high-risk scores exhibited significantly worse OS, lower numbers of TILs, and lower sensitivity to chemotherapy drugs than patients with low-risk scores.
Conclusions: Overall, we successfully established a novel prognostic model based on ARLncs, which holds significant promise for predicting prognosis and personalized therapy for patients with HNSCC.
Keywords: Anoikis; head and neck squamous cell carcinoma (HNSCC); long non-coding RNA (lncRNA); overall survival (OS); prognosis.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2520/coif). The authors have no conflicts of interest to declare.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
