Development of a quantitative genomic instability scoring system and a related competing endogenous RNA network in head and neck squamous cell carcinoma
- PMID: 40792171
- PMCID: PMC12335705
- DOI: 10.21037/tcr-24-1925
Development of a quantitative genomic instability scoring system and a related competing endogenous RNA network in head and neck squamous cell carcinoma
Abstract
Background: Genomic instability (GI) is a hallmark of cancer and plays a crucial role in the progression of head and neck squamous cell carcinoma (HNSCC). This study aimed to quantitatively characterize GI features and construct a GI-related competing endogenous RNA (ceRNA) network in HNSCC.
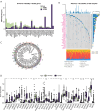
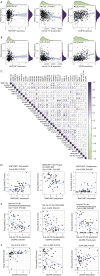
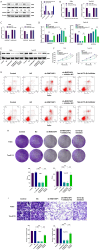
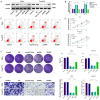
Methods: Weighted gene co-expression network analysis (WGCNA) and differential gene expression analysis were conducted to compare genomically stable and unstable HNSCC samples. Thirty-six hub GI-related genes (GIGs) were identified and used to categorize patients into distinct clusters through consensus clustering analysis. A GI scoring (GIS) system was then developed to assess its relationship with somatic mutations, tumor mutational burden (TMB), and differential gene expression, including genes such as KRAS and TP53. In vitro experiments were performed to explore the functional mechanism of the GI-associated ceRNA axis-RNF216P1/let-7b-5p/DUSP9. The expression levels of RNF216P1, let-7b-5p, and DUSP9 were also validated using clinical samples from a local hospital.
Results: The identified 36 GIGs enabled the categorization of HNSCC patients into three distinct clusters, each exhibiting unique prognostic and immune profiles. The developed GIS system effectively distinguished between somatic mutations, TMB, and differential gene expression. Patients with higher GIS scores had better prognoses compared to those with lower scores. Additionally, GIS was positively correlated with overall immune cell infiltration and immune function, highlighting its potential in predicting responses to immunotherapy. The GI-associated ceRNA axis RNF216P1/let-7b-5p/DUSP9 was established, with The Cancer Genome Atlas (TCGA) analysis revealing upregulation of RNF216P1 and DUSP9 in tumor tissues, while let-7b-5p was downregulated. These expression trends were corroborated in clinical samples. In vitro experiments demonstrated that RNF216P1 functioned as a molecular sponge for let-7b-5p, leading to upregulation of DUSP9 and promoting oncogenesis in HNSCC.
Conclusions: The GIS system is an effective biomarker for evaluating GI, prognosis, and immune features in HNSCC. The findings also clarify the functional mechanism of the GI-related ceRNA axis RNF216P1/let-7b-5p/DUSP9, providing valuable insights for future research and the development of therapeutic strategies for HNSCC.
Keywords: Genomic instability (GI); competing endogenous RNA (ceRNA); head and neck squamous cell carcinoma (HNSCC); immunity.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1925/coif). The authors have no conflicts of interest to declare.
Figures








References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
