Exploring the biological basis for the identification of different syndromes in ischemic heart failure based on joint multi-omics analysis
- PMID: 40792212
- PMCID: PMC12337011
- DOI: 10.3389/fphar.2025.1641422
Exploring the biological basis for the identification of different syndromes in ischemic heart failure based on joint multi-omics analysis
Abstract
Background: IHF is a major chronic disease that seriously threatens human health. Qi deficiency and blood stasis syndrome (QDBS), Yang deficiency with blood stasis syndrome (YDBS) and Yang deficiency and blood stasis with fluid retention syndrome (YDBSFR) are the basic syndromes of IHF in Chinese medicine. This study aims to explore the biological basis of the three IHF syndromes through integrated multi-omics research.
Methods: We analyzed and integrated transcriptomic, proteomic, and targeted metabolomic data from IHF patients and healthy persons to obtain the key biomarkers and enriched pathways of QDBS, YDBS and YDBSFR(Registration No.: ChiCTR2200058314). These biomarkers were combined with clinical indicators to construct the "Disease-Syndromes-Clinical phenotypes-Biomarkers-Pathways" network, and the obtained differential genes and proteins were externally validated.
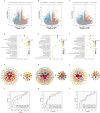
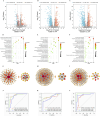
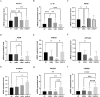
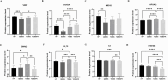
Results: The potential biomarkers for QDBS included SDHD, IL10, ACTG1, VWF, MDH2, COX5A, Valeric acid, Succinic Acid and L-Histidine, which were predominantly enriched in TCA cycle, oxidative phosphorylation, platelet activation, and neutrophil extracellular trap formation pathways, demonstrating associations with energy metabolism, coagulation system, and immune-inflammatory responses.YDBS potential biomarkers included TSHR, PRKG1, ATP1A2, GNAI2, APOA2, PLTP, 3-Hydroxybutyrate, Hexadecanoic acid and Palmitelaidic acid, and the combined pathways were mainly enriched in thyroid hormone synthesis, regulation of lipolysis in adipocytes, cholesterol metabolism and PPAR signaling pathways, correlating with hormonal regulation and lipid metabolism. The potential biomarkers of YDBSFR were CNGB1, KCNMA1, PIK3R2, HSPA8, C3, FH, Oxamic acid, N-Acetyl-L-alanine, 4-Hydroxyhippuric acid, and the combined pathways were mainly enriched in aldosterone-regulated sodium reabsorption, cGMP-PKG signaling pathway, neutrophil extracellular trap formation and TCA cycle signaling pathways, which are related to hormone regulation, signal transduction, immune-inflammatory response and energy metabolism. Platelet activation was involved in the whole process of IHF. External validation demonstrated the above core targets.
Conclusion: This study investigated the biological basis of QDBS, YDBS and YDBSFR in IHF from a modern biomedical perspective, providing references for the objective research of TCM syndrome differentiation.
Keywords: biological basis; ischemic heart failure; multi-omics; syndrome differentiation; traditional Chinese medicine.
Copyright © 2025 Zhang, Wei, Qiao, Yu, Ren, Zhao, Sun, Wang, Li, Wang and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Integrating multi-omics and machine learning strategies to explore the "gene-protein-metabolite" network in ischemic heart failure with Qi deficiency and blood stasis syndrome.Chin Med. 2025 Jul 17;20(1):93. doi: 10.1186/s13020-025-01151-9. Chin Med. 2025. PMID: 40671145 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Wenjing decoction exerts analgesic effects on cold coagulation and stasis PD through BDNF/ TRKB/CREB pathway.J Ethnopharmacol. 2025 Jul 3;352:120220. doi: 10.1016/j.jep.2025.120220. Online ahead of print. J Ethnopharmacol. 2025. PMID: 40617361
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
-
Blood biomarkers for the non-invasive diagnosis of endometriosis.Cochrane Database Syst Rev. 2016 May 1;2016(5):CD012179. doi: 10.1002/14651858.CD012179. Cochrane Database Syst Rev. 2016. PMID: 27132058 Free PMC article.
References
-
- Bano A., Chaker L., Muka T., Mattace-Raso F. U. S., Bally L., Franco O. H., et al. (2020). Thyroid function and the risk of fibrosis of the liver, heart, and lung in humans: a systematic review and meta-analysis. Thyroid official J. Am. Thyroid Assoc. 30, 806–820. 10.1089/thy.2019.0572 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

