Tomato spotted wilt virus in tomato from Croatia, Montenegro and Slovenia: genetic diversity and evolution
- PMID: 40792267
- PMCID: PMC12336143
- DOI: 10.3389/fmicb.2025.1618327
Tomato spotted wilt virus in tomato from Croatia, Montenegro and Slovenia: genetic diversity and evolution
Abstract
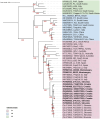
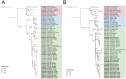
Tomato spotted wilt orthotospovirus (TSWV) is a major plant pathogen causing significant economic losses in tomato production worldwide. Understanding its genetic diversity and evolutionary mechanisms is crucial for effective disease management. This study analyzed TSWV isolates from symptomatic tomato plants collected across Croatia, Montenegro and Slovenia between 2020 and 2024. High-throughput sequencing (HTS) was employed to obtain whole-genome sequences, followed by phylogenetic analyses to assess genetic variability and relationships among isolates from these three countries and other isolates of worldwide geographic origin. Phylogenetic analyses placed all studied isolates within the L1-M3-S3 genotype, commonly associated with solanaceous crops in Europe. While Croatian and Slovenian isolates exhibited high genetic similarity, Montenegrin isolates clustered in a distinct subgroup, showing closer relationships to Asian and Mediterranean accessions. Despite the severe disease symptoms observed, no substitutions in the NSm protein associated with resistance-breaking (RB) phenotypes were detected. These findings suggest that additional virome components, environmental factors or so far unknown mechanism(s) may contribute to infection and disease severity in tomato and strongly support the need of continuous surveillance of TSWV genetic diversity in order to inform breeding programs and develop sustainable management strategies to mitigate future outbreaks.
Keywords: HTS; TSWV; phylogeny; plant virus; tomato.
Copyright © 2025 Škorić, Zindović, Grbin, Pul, Božović, Margaria, Mehle, Pecman, Kogej Zwitter, Kutnjak and Vučurović.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
An all-out assault on a dominant resistance gene: Local emergence, establishment, and spread of strains of tomato spotted wilt orthotospovirus (TSWV) that overcome Sw-5b-mediated resistance in fresh market and processing tomatoes in California.PLoS One. 2024 Jul 10;19(7):e0305402. doi: 10.1371/journal.pone.0305402. eCollection 2024. PLoS One. 2024. PMID: 38985801 Free PMC article.
-
Insights into the genetic variability and evolutionary dynamics of tomato spotted wilt orthotospovirus in China.BMC Genomics. 2024 Jan 8;25(1):40. doi: 10.1186/s12864-023-09951-9. BMC Genomics. 2024. PMID: 38191299 Free PMC article.
-
Molecular genetic analyses of the N, NSm and NSs genes of a local population of Orthotospovirus tomatomaculae reveal purifying selection in crops in the southeastern USA.J Gen Virol. 2025 Jul;106(7):002119. doi: 10.1099/jgv.0.002119. J Gen Virol. 2025. PMID: 40622855 Free PMC article.
-
Natural Resources Resistance to Tomato Spotted Wilt Virus (TSWV) in Tomato (Solanum lycopersicum).Int J Mol Sci. 2021 Oct 12;22(20):10978. doi: 10.3390/ijms222010978. Int J Mol Sci. 2021. PMID: 34681638 Free PMC article. Review.
-
The clinical effectiveness and cost-effectiveness of enzyme replacement therapy for Gaucher's disease: a systematic review.Health Technol Assess. 2006 Jul;10(24):iii-iv, ix-136. doi: 10.3310/hta10240. Health Technol Assess. 2006. PMID: 16796930
References
-
- Adkins S., Turina M., Whitfield A., Resende R., Naidu R., Hughes H. (2022). Rename all existing species to comply with the binomial species format (Bunyavirales: Tospoviridae). Available online at: https://ictv.global/ictv/proposals/2022.011P.Tospoviridae_rename.zip.
-
- Almási A., Pinczés D., Tímár Z., Sáray R., Palotás G., Salánki K. (2023). Identification of a new type of resistance breaking strain of tomato spotted wilt virus on tomato bearing the Sw-5b resistance gene. Eur. J. Plant Pathol. 166, 219–225. doi: 10.1007/s10658-023-02656-5 - DOI
-
- Batuman O., Turini T. A., LeStrange M., Stoddard S., Miyao G., Aegerter B. J., et al. (2020). Development of an IPM strategy for Thrips and tomato spotted wilt virus in processing tomatoes in the Central Valley of California. Pathogens 9:636. doi: 10.3390/pathogens9080636, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

