The Transcription Factor CREB1 Triggers the Progression of Clear Cell Renal Cell Carcinoma by Promoting CENPE Expression
- PMID: 40794013
- PMCID: PMC12341428
- DOI: 10.1111/jcmm.70773
The Transcription Factor CREB1 Triggers the Progression of Clear Cell Renal Cell Carcinoma by Promoting CENPE Expression
Abstract
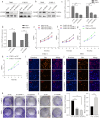
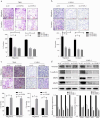
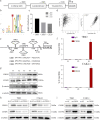
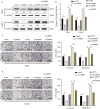
Centromere-associated protein E (CENPE) has been identified as overexpressed in multiple cancers and exerts a tumour promotion function by affecting chromosome misalignment and mitosis. However, the expression pattern, biological roles, and underlying molecular mechanism of CENPE in clear cell renal cell carcinoma (ccRCC) progression have not been fully elucidated. In the present study, the expression levels of CENPE in ccRCC and paracancerous specimens were measured using the public RNA sequencing data and validated in a cohort of ccRCC samples from our centre. We found that CENPE was significantly over-expressed in ccRCC tissues and promoted proliferative and metastatic abilities of ccRCC cells and xenografts through regulating the epithelial-mesenchymal transition (EMT) process. Furthermore, bioinformatic analysis and ChIP assay indicated that the transcription factor CREB1 bound to the promoter region of CENPE and activated its transcription in ccRCC cells. Taken together, our findings demonstrated that the CREB1-CENPE axis was responsible for stimulating the in vitro and in vivo progression of ccRCC, serving as a promising therapeutic target for ccRCC.
Keywords: CENPE; CREB1; EMT; ccRCC; progression.
© 2025 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

