Design and characterization of HIV-1 vaccine candidates to elicit antibodies targeting multiple epitopes
- PMID: 40794027
- PMCID: PMC12341506
- DOI: 10.1084/jem.20250693
Design and characterization of HIV-1 vaccine candidates to elicit antibodies targeting multiple epitopes
Abstract
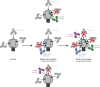
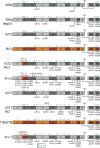
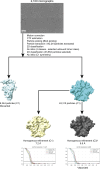
A primary goal in the development of an AIDS vaccine is the elicitation of broadly neutralizing antibodies (bNAbs) that protect against diverse HIV-1 strains. To this aim, germline-targeting immunogens have been developed to activate bNAb precursors and initiate the induction of bNAbs. While most preclinical germline-targeting HIV-1 vaccine candidates only include a single bNAb precursor epitope, an effective HIV-1 vaccine will likely require bNAbs that target multiple epitopes on Env. Here, we report a newly designed germline-targeting Env SOSIP trimer, named 3nv.2, that presents three bNAb epitopes on Env: the CD4bs, V3, and V2 epitopes. 3nv.2 forms a stable trimeric Env and binds to bNAb precursors from each of the desired epitopes. Immunization experiments in rhesus macaques and mice demonstrate 3nv.2 elicits the combined effects of its parent immunogens. Our results provide proof of concept for using a germline-targeting immunogen presenting three or more bNAb epitopes and a framework to develop improved next-generation HIV-1 vaccine candidates.
© 2025 Gristick et al.
Conflict of interest statement
Disclosures: H.B. Gristick reported a patent to PCT/US2023/068921 pending. H. Hartweger reported a patent to HIV Vaccine Immunogens WO2023250448A1 pending. M.C. Nussenzweig reported personal fees from Celldex Pharmaceutical outside the submitted work. P.J. Bjorkman reported a patent to PCT/US2023/068921 pending. No other disclosures were reported.
Figures



Update of
-
Design and characterization of HIV-1 vaccine candidates to elicit antibodies targeting multiple epitopes.bioRxiv [Preprint]. 2025 Jul 21:2025.01.08.632013. doi: 10.1101/2025.01.08.632013. bioRxiv. 2025. Update in: J Exp Med. 2025 Oct 6;222(10):e20250693. doi: 10.1084/jem.20250693. PMID: 39829910 Free PMC article. Updated. Preprint.
References
-
- Barnes, C.O., Gristick H.B., Freund N.T., Escolano A., Lyubimov A.Y., Hartweger H., West A.P. Jr., Cohen A.E., Nussenzweig M.C., and Bjorkman P.J.. 2018. Structural characterization of a highly-potent V3-glycan broadly neutralizing antibody bound to natively-glycosylated HIV-1 envelope. Nat. Commun. 9:1251. 10.1038/s41467-018-03632-y - DOI - PMC - PubMed
-
- Bonsignori, M., Hwang K.-K., Chen X., Tsao C.-Y., Morris L., Gray E., Marshall D.J., Crump J.A., Kapiga S.H., Sam N.E., et al. 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85:9998–10009. 10.1128/JVI.05045-11 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

