Deep learning-based radiolabelled compound-protein interaction prediction for NDUFS1-targeting radiopharmaceutical discovery
- PMID: 40794258
- PMCID: PMC12343451
- DOI: 10.1186/s13550-025-01300-z
Deep learning-based radiolabelled compound-protein interaction prediction for NDUFS1-targeting radiopharmaceutical discovery
Abstract
Background: NDUFS1 is the largest subunit of OXPHOS complex I (MC-I) and mutations in this gene are associated with MC-I deficiency. This study aims to develop a graph neural network and attention mechanism-based radiopharmaceutical-protein (RP-protein) interaction prediction model for identifying an imaging candidate of mitochondrial function through targeting its core subunit NDUFS1.
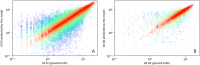
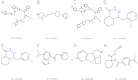
Results: The estimated cell viability values for trastuzumab, 177Lu-DOTA-trastuzumab, and 225Ac-DOTA-trastuzumab were 290.1, 89.01, and 8.262 nM, respectively. The deep learning (DL) model was pretrained with normal compound-protein pairs. Afterwards, the model was fine-tuned with the dataset of RP-protein pairs and evaluated with five-fold cross validation. The prediction model trained with normal compound-protein pairs effectively predicted the binding affinity. The fine-tuned model incorporating radioactive properties outperformed the same model trained only on normal compounds. The model estimated the important substructure of a compound related to its binding to the target protein. NDUFS1 protein-targeting compounds were identified and BDBM210829 compound had the best binding affinities, binding rank, and LogP as it binds to the NDUFS1.
Conclusions: This study proposed a DL-based radiolabelled compound-protein interaction prediction model to identify a radiopharmaceutical (RP) that binds to the mitochondrial core subunit NDUFS1. The proposed model shows good performance for predicting RP-protein interaction. BDBM210829 was identified as a top candidate for radiolabeling and targeting the mitochondrial core subunit NDUFS1. This model can be used as an effective virtual screening tool for RP discovery.
Supplementary Information: The online version contains supplementary material available at 10.1186/s13550-025-01300-z.
Keywords: Binding affinity; Compound protein interaction; Graph neural network; Mitochondria; NDUFS1; Radiopharmaceutical discovery.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the Institutional Review Board of KIRAMS (IRB No.: 2022-09-006-002). All methods were performed in accordance with the relevant guidelines and regulations. Consent for publication: Informed consent was obtained from all participants involved in the study. Competing interests: The authors have declared that no competing interest exists. Clinical trial number: Not applicable.
Figures


Similar articles
-
Improved compound-protein interaction site and binding affinity prediction using self-supervised protein embeddings.BMC Bioinformatics. 2022 Dec 16;23(1):543. doi: 10.1186/s12859-022-05107-w. BMC Bioinformatics. 2022. PMID: 36526969 Free PMC article.
-
DMHGNN: Double multi-view heterogeneous graph neural network framework for drug-target interaction prediction.Artif Intell Med. 2025 Jan;159:103023. doi: 10.1016/j.artmed.2024.103023. Epub 2024 Nov 17. Artif Intell Med. 2025. PMID: 39579417
-
A deep learning method for drug-target affinity prediction based on sequence interaction information mining.PeerJ. 2023 Dec 11;11:e16625. doi: 10.7717/peerj.16625. eCollection 2023. PeerJ. 2023. PMID: 38099302 Free PMC article.
-
Drug-drug interaction analysis based on information bottleneck graph neural network: A review.Medicine (Baltimore). 2025 Jun 20;104(25):e42904. doi: 10.1097/MD.0000000000042904. Medicine (Baltimore). 2025. PMID: 40550083 Free PMC article. Review.
-
Prediction of drug-target binding affinity based on deep learning models.Comput Biol Med. 2024 May;174:108435. doi: 10.1016/j.compbiomed.2024.108435. Epub 2024 Apr 8. Comput Biol Med. 2024. PMID: 38608327 Review.
References
-
- Chhikara BS, Parang K. Global cancer statistics 2022: the trends projection analysis. Chem Biol Lett. 2023;10(1):451.
-
- Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK, Howell A, et al. Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: visualizing the therapeutic effects of Metformin in tumor tissue. Cell Cycle. 2011;10(23):4047–64. 10.4161/cc.10.23.18151. - PMC - PubMed
-
- Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta. 2012;1817(6):851–62. 10.1016/j.bbabio.2011.08.010. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

