Blood metabolomics improves prediction of central nervous system damage in multiple sclerosis
- PMID: 40794313
- PMCID: PMC12343719
- DOI: 10.1007/s11306-025-02315-2
Blood metabolomics improves prediction of central nervous system damage in multiple sclerosis
Abstract
Introduction: Multiple sclerosis (MS) is an autoimmune disorder with an unpredictable outcome at the time of diagnosis. The measurement of serum neurofilament light chain (sNfL) and glial fibrillary acidic protein (sGFAP) has introduced new biomarkers for assessing MS disease activity and progression. However, there is a need for additional diagnostic and prognostic tools. In this study, we investigated the predictive abilities of metabolomics, gut microbiota, as well as clinical and lifestyle factors for MS outcome parameters.
Objectives: The aim of this study was to assess the predictive capacity of plasma metabolites, gut microbiota, and clinical/lifestyle factors on MS outcome measures including MS-related fatigue, MS disability, and sNfL and sGFAP concentrations.
Methods: A prospective cohort study was conducted with 54 individuals with MS. Anthropometric, biological, and lifestyle parameters were collected. The least absolute shrinkage and selection operator (LASSO) algorithm with ten-fold cross-validation was used to identify predictors of MS disease outcome parameters based on plasma metabolomics, microbiota sequencing, and clinical and lifestyle measurements obtained from questionnaires and anthropometric measurements.
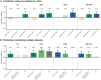
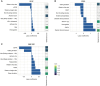
Results: Circulating metabolites were found to be superior predictors for sNfL and sGFAP concentrations, while clinical and lifestyle data were associated with EDSS scores. Both plasma metabolites and clinical data significantly predicted MS-related fatigue. Combining multiple multi-omics data did not consistently improve predictive performance.
Conclusions: This study highlights the value of plasma metabolites as predictors of sNfL, sGFAP, and fatigue in MS. Our findings suggest that prioritizing metabolomics over other methods can lead to more accurate predictions of MS disease outcomes.
Keywords: Biomarkers; Gut-microbiota; Metabolomics; Multiple sclerosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: All authors declare that they have no conflict of interest. Ethical approval: Each participant signed a written informed consent form approved by the local Ethics Committee (CER-VD 2018–01862) before inclusion. All procedures involving human participants were conducted following the ethical standards of the institutional and/or national research committee and relevant ethical guidelines.
Figures


References
-
- Apetoh, L., Quintana, F. J., Pot, C., Joller, N., Xiao, S., Kumar, D., Burns, E. J., Sherr, D. H., Weiner, H. L., & Kuchroo, V. K. (2010). The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nature Immunology,11(9), 854–861. 10.1038/ni.1912 - PMC - PubMed
-
- Benkert, P., Meier, S., Schaedelin, S., Manouchehrinia, A., Yaldizli, Ö., Maceski, A., Oechtering, J., Achtnichts, L., Conen, D., Derfuss, T., & Lalive, P. H. (2022). Serum neurofilament light chain for individual prognostication of disease activity in people with multiple sclerosis: A retrospective modelling and validation study. The Lancet Neurology,21(3), 246–257. 10.1016/S1474-4422(22)00009-6 - PubMed
-
- Berer, K., Gerdes, L. A., Cekanaviciute, E., Jia, X., Xiao, L., Xia, Z., Liu, C., Klotz, L., Stauffer, U., Baranzini, S. E., & Kümpfel, T. (2017). Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proceedings of the National Academy of Sciences of the United States of America,114(40), 10719–10724. 10.1073/pnas.1711233114 - PMC - PubMed
-
- Buysse, D. J., Reynolds, C. F., Monk, T. H., Berman, S. R., & Kupfer, D. J. (1989). The Pittsburgh sleep quality index: A new instrument for psychiatric practice and research. Psychiatry Research,28(2), 193–213. 10.1016/0165-1781(89)90047-4 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
