Divergent TIR signaling domains in TLR7 and TLR9 control opposing effects on systemic autoimmunity
- PMID: 40794441
- PMCID: PMC12578402
- DOI: 10.1172/JCI189566
Divergent TIR signaling domains in TLR7 and TLR9 control opposing effects on systemic autoimmunity
Abstract
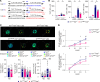
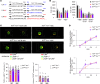
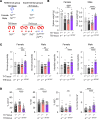
Toll like receptor (TLR) 7 and 9, endosomal sensors for RNA and DNA, are key mediators of autoreactivity. Although generally considered homologous, they paradoxically have opposing effects on lupus: TLR7 exacerbates the disease while TLR9 protects from it How they mediate opposing effects in autoimmunity remains undetermined. We hypothesized that differences in signaling qualities of the Toll-Interleukin 1 Receptor (TIR) domains of TLR7 and TLR9 could be responsible for their opposing effects. To test this, we introduced the TIR domain of TLR9 into the endogenous Tlr7 locus and the TLR7 TIR domain into the endogenous Tlr9 locus of mice, creating chimeric molecules termed TLR779 and TLR997. Lupus-prone MRL/lpr mice carrying Tlr779 had greatly ameliorated disease, while MRL/lpr mice carrying Tlr997 had markedly exacerbated disease compared with respective TlrWT mice. These experiments establish that TLR7 and TLR9 TIR domains have divergent properties and control disease quality, thus explaining the longstanding "TLR paradox".
Keywords: Autoimmunity; Immunology; Innate immunity; Lupus.
Conflict of interest statement
Figures




Comment in
- S-TIR-ring up TLR7 and TLR9: signaling domain substitutions clarify the TLR paradox
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

