Spatial proteomics reveals signal sequence characteristics correlated with localization in cyanobacteria
- PMID: 40794799
- PMCID: PMC12341915
- DOI: 10.1093/plphys/kiaf186
Spatial proteomics reveals signal sequence characteristics correlated with localization in cyanobacteria
Abstract
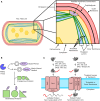
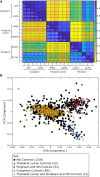
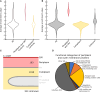
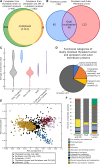
Cyanobacteria have an inner and outer cell membrane enclosing the periplasm and cell wall and an additional set of internal membranes (called the thylakoid membranes) enclosing the thylakoid lumen. The periplasm and thylakoid lumen have unique proteomes, but the mechanisms regulating protein sorting to these locations have remained elusive. Here, proximity-based proteomics using the engineered peroxidase APEX2 was performed in the cyanobacteria Synechococcus sp. PCC 7002 to profile the proteomes of the cytoplasm, thylakoid lumen, and the periplasm and outer membrane (P-OM). Our analyses revealed specific roles for the thylakoid lumen in photosynthesis and energy generation, as well as roles for the periplasm in metabolite transport and binding, cell motility, and cell wall maintenance. Forty proteins localized to both the thylakoid lumen and the P-OM; however, their biological functions remain unclear. We also analyzed the correlation between signal sequence characteristics and differential protein localization to either the thylakoid lumen or the P-OM. In PCC 7002, as well as Synechocystis sp. PCC 6803 and Nostoc sp. PCC 7120, thylakoid lumen proteins translocated across membranes via the Secretory (Sec) system possessed more hydrophobic and alpha-helical signal sequence H-regions than P-OM proteins. The signal sequences of homologous proteins in Gloeobacter violaceus PCC 7421, a cyanobacterial species with a combined thylakoid lumen and periplasmic space, did not exhibit such differences. Therefore, the pattern of increased H-region hydrophobicity and alpha helix content is specific to cyanobacteria with a separate thylakoid lumen space and likely contributes to proper protein sorting between the thylakoid lumen and periplasm.
© The Author(s) 2025. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

