All-in-one medical image-to-image translation
- PMID: 40795870
- PMCID: PMC12461644
- DOI: 10.1016/j.crmeth.2025.101138
All-in-one medical image-to-image translation
Abstract
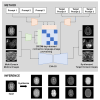
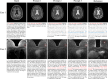
The growing availability of public multi-domain medical image datasets enables training omnipotent image-to-image (I2I) translation models. However, integrating diverse protocols poses challenges in domain encoding and scalability. Therefore, we propose the "every domain all at once" I2I (EVA-I2I) translation model using DICOM-tag-informed contrastive language-image pre-training (DCLIP). DCLIP maps natural language scan descriptions into a common latent space, offering richer representations than traditional one-hot encoding. We develop the model using seven public datasets with 27,950 scans (3D volumes) for the brain, breast, abdomen, and pelvis. Experimental results show that our EVA-I2I can synthesize every seen domain at once with a single training session and achieve excellent image quality on different I2I translation tasks. Results for downstream applications (e.g., registration, classification, and segmentation) demonstrate that EVA-I2I can be directly applied to domain adaptation on external datasets without fine-tuning and that it also enables the potential for zero-shot domain adaptation for never-before-seen domains.
Keywords: CP: Computational biology; CP: Imaging; contrastive language-image pre-training; image-to-image translation; multi-domain medical image; representation learning; zero-shot domain adaptation.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Pooch E.H., Ballester P., Barros R.C. Thoracic Image Analysis: Second International Workshop, TIA 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 8, 2020, Proceedings 2. Springer; 2020. Can we trust deep learning based diagnosis? the impact of domain shift in chest radiograph classification; pp. 74–83.
-
- Torralba A., Efros A.A. CVPR 2011. IEEE; 2011. Unbiased look at dataset bias; pp. 1521–1528.
-
- Perone C.S., Ballester P., Barros R.C., Cohen-Adad J. Unsupervised domain adaptation for medical imaging segmentation with self-ensembling. Neuroimage. 2019;194:1–11. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

