The Influence of CG sites on dynamic DNA sequence mutagenesis in the genomic evolution of mammalian lifespan
- PMID: 40795962
- PMCID: PMC12342937
- DOI: 10.1093/nar/gkaf762
The Influence of CG sites on dynamic DNA sequence mutagenesis in the genomic evolution of mammalian lifespan
Abstract
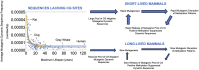
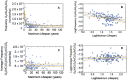
Previous work showed that natural selection has acted to minimize the genomic frequencies of representative dynamic DNA sequences capable of forming G-quadruplex, Triplex, hairpin, and i-motif structures in long-lived mammals, thus diminishing the mutagenic potential of their genomes. This report extends findings with single sequences to broadly distributed G3-4N1-7G3-4N1-7G3-4N1-7G3-4 dynamic sequence motifs and identifies a second, previously unknown, pool of dynamic DNA sequences that escape negative selective pressure as a function of lifespan. This pool is distinguished from those studied previously by the presence of one or more CG sites, suggesting that they are subject to structural suppression DNA methylation in mammals. Consistent with the known effects of DNA damage on methylation patterns, the frequencies of dynamic sequences that lack CG sites were found to track species-specific mutation rate and species-specific methylation rates in 126 genomes representing 26 mammalian orders. The results suggest that DNA methylation itself and perhaps methylated DNA binding proteins also function in the suppression of the mutagenic potential of dynamic sequences containing CG sites, and that this latent pool of mutagenic potential is released during the mutation induced decay of DNA methylation patterns linked to the inborn level of dynamic sequences lacking CG sites.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
The author has no conflicts to interest to disclose.
Figures








References
MeSH terms
LinkOut - more resources
Full Text Sources

