Stratifying IVF population endometria using a prognosis gradient independent of endometrial timing†
- PMID: 40796355
- PMCID: PMC12491665
- DOI: 10.1093/humrep/deaf156
Stratifying IVF population endometria using a prognosis gradient independent of endometrial timing†
Abstract
Study question: Can the disrupted window of implantation (WOI) be stratified according to transcriptomic patterns associated with reproductive success in IVF patients undergoing HRT?
Summary answer: There are four transcriptomic patterns independent of endometrial timing associated with a gradient of reproductive prognosis underlying different molecular pathomechanisms.
What is known already: A molecular heterogeneous profile independent of endometrial timing has been discovered as a cause of implantation failure that disrupt the endometrial transcriptome in the mid-secretory phase. However, the molecular heterogeneous patterns underlying the disruption remain poorly identify and understood. Characterizing the molecular heterogeneity of this endometrial disruption is crucial to develop personalized and more accurate diagnostic tools for preventive medicine, particularly for patients with a high risk of endometrial failure.
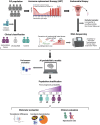
Study design, size, duration: In this multicenter prospective study, 195 IVF patients undergoing HRT with endometrial biopsy collection, during mid-secretory phase for endometrial progression evaluation, were recruited between January 2019 and August 2022. Out of 195 patients, 131 were finally included in the following analysis.
Participants/materials, setting, methods: Endometrial biopsies were processed for whole endometrial transcriptome analysis using RNA-Sequencing. To identify disruptions in the WOI, the transcriptomic variation due to cyclic endometrial tissue changes was removed. Out of 195 biopsies sequenced, 131 were derived from patients that met the clinical criteria to be classified as implantation failure group (≥3 implantation failures, n = 32) or control group (<3 implantation failures, n = 99). An artificial intelligence (AI) model, based on two supervised learning algorithms: support vector machine (SVM) and k-nearest neighbors (kNN), was performed with 131 patients that were randomly allocated to training (n = 105) and test (n = 26) sets for biomarker signature discovery and assessment of predictive performance, respectively. The reproductive outcomes of the single embryo transfer immediately after biopsy collection were analyzed. Differential expression and functional analyses were performed to characterize molecular profiles. Finally, a quantitative PCR (qPCR) assay was used to corroborate the differential expression of six potential biomarkers.
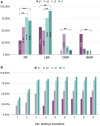
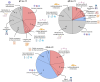
Main results and the role of chance: With the dichotomous clinical classification of poor or good reproductive prognosis, there was no transcriptomic distinction between patients with a history of implantation failures during HRT endometrial preparation. Alternatively, using an AI model to stratify IVF patients based on the probability of endometrial disruption revealed molecular and clinical differences between patterns. Patients were stratified into four reproductive prognosis-related profiles: p1 (n = 24), p2 (n = 14), c2 (n = 32) and c1 (n = 61). The highest pregnancy rate (PR) was associated with c1 (91%) and the highest ongoing pregnancy rate (OPR) was associated with c2 (78%), linking these profiles to good reproductive prognoses. On the other hand, p1 had the highest biochemical miscarriage rate (43%) while p2 had the highest clinical miscarriage rate (43%). Notably, both p1 and p2 were related to lower PR and OPR, supporting that these profiles were associated with poor prognoses. Regarding the functional characterization in the poor prognosis profiles that were linked to miscarriages, p1 was associated with an excessive immune response against the embryo during early pregnancy stages, while p2 was initially immune-tolerant but rejected the fetus in later stages due to the lack of metabolic response.
Limitations, reasons for caution: Due to the heterogeneous character of the disrupted WOI and the limited sample size of the different stratified groups, the AI model has limited population inference. However, our significant promising findings provide strong leads for further clinical studies with larger sample sizes.
Wider implications of the findings: This new transcriptomic taxonomy associated with distinct reproductive outcomes provides clues to design new and more accurate evaluation tools for endometrial-factor infertility. Furthermore, it enables tailoring therapeutic strategies to apply a personalized medicine to each patient suffering from endometrial-factor infertility, improving their odds of getting pregnant.
Study funding/competing interest(s): This study was supported by the IVI Foundation (1706-FIVI-048-PD); Instituto de Salud Carlos III (ISCIII) and co-funded by the European Regional Development Fund "A way to make Europe" (PI19/00537 [P.D.-G.]) as well as Instituto Carlos III (ISCIII) through project (PI23/00806 [P.D.-G.]) and co-funded by European Union. Patricia Diaz-Gimeno is supported by Instituto de Salud Carlos III (ISCIII) through the Miguel Servet program (CP20/00118) co-funded by the European Union. Patricia Sebastian-Leon and Francisco Jose Sanz are funded by Instituto de Salud Carlos III (ISCIII) through the Sara Borrell postdoctoral program (CD21/00132 [P.S.-L.] and CD23/00032 [F.J.S.]) co-financed by the European Union. Josefa Maria Sanchez-Reyes was supported by a predoctoral fellowship program of the Generalitat Valenciana (ACIF/2018/072 and BEFPI/2020/028). Antonio Parraga-Leo (FPU18/01777) and Diana Marti-Garcia (FPU19/03247) were supported by predoctoral fellowship programs of the Spanish Ministry of Science, Innovation and Universities. The authors declare no conflicts of interest.
Trial registration number: Not applicable.
Keywords: artificial intelligence; endometrial disruption; endometrial function; endometrial transcriptomics; gene expression signature; infertility; precision medicine; transcriptomic stratification.
© The Author(s) 2025. Published by Oxford University Press on behalf of European Society of Human Reproduction and Embryology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Andrews S, Krueger F, Segonds-Pichon A, Biggins L, Krueger C, Wingett S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2012. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (15 July 2024, date last accessed).
-
- Bell J. Stratified medicines: towards better treatment for disease. Lancet 2014;383:S3–S5. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

