REST/NRSF Preserves muscle stem cell identity by repressing alternate cell fate
- PMID: 40796549
- PMCID: PMC12343973
- DOI: 10.1038/s41467-025-62758-y
REST/NRSF Preserves muscle stem cell identity by repressing alternate cell fate
Abstract
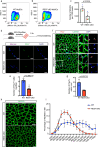
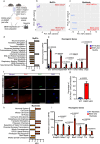
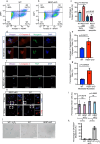
Cell fate and identity require timely activation of lineage-specific and concomitant repression of alternate-lineage genes. How this process is epigenetically encoded remains largely unknown. In skeletal muscle stem cells, the myogenic regulatory factors are well-established drivers of muscle gene activation but less is known about how non-muscle gene repression is achieved. Here, we show that the master epigenetic regulator, Repressor Element 1-Silencing Transcription factor (REST), also known as Neuron-Restrictive Silencer Factor (NRSF), is a key regulator of this process. We show that many non-lineage genes retain permissive chromatin state but are actively repressed by REST. Loss of functional REST in muscle stem cells and progenitors disrupts muscle specific epigenetic and transcriptional signatures, impairs differentiation, and triggers apoptosis in progenitor cells, leading to depletion of the stem cell pool. Consequently, REST-deficient skeletal muscle exhibits impaired regeneration and reduced myofiber growth postnatally. Collectively, our data suggests that REST plays a key role in safeguarding muscle stem cell identity by repressing multiple non-muscle lineage and developmentally regulated genes in adult mice.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Wang, Y. X. & Rudnicki, M. A. Satellite cells, the engines of muscle repair. Nat. Rev. Mol. Cell Biol.13, 127–133 (2011). - PubMed
-
- Schultz, E., Gibson, M. C. & Champion, T. Satellite cells are mitotically quiescent in mature mouse muscle: an EM and radioautographic study. J. Exp. Zool.206, 451–456 (1978). - PubMed
-
- Collins, C. A. et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell122, 289–301 (2005). - PubMed
-
- Maltin, C. A., Harris, J. B. & Cullen, M. J. Regeneration of mammalian skeletal muscle following the injection of the snake-venom toxin, taipoxin. Cell Tissue Res.232, 565–577 (1983). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

