Modeling the genomic architecture of adiposity and anthropometrics across the lifespan
- PMID: 40796553
- PMCID: PMC12343819
- DOI: 10.1038/s41467-025-62730-w
Modeling the genomic architecture of adiposity and anthropometrics across the lifespan
Abstract
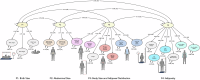
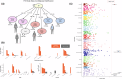
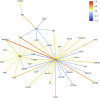
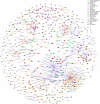
Obesity-related conditions are among the leading causes of preventable death and are increasing in prevalence worldwide. Body size and composition are complex traits that are challenging to characterize due to environmental and genetic influences, longitudinal variation, heterogeneity between sexes, and differing health risks based on adipose distribution. Here, we construct a 4-factor genomic structural equation model using 18 measures, unveiling shared and distinct genetic architectures underlying birth size, abdominal size, adipose distribution, and adiposity. Multivariate genome-wide associations reveal the adiposity factor is enriched specifically in neural tissues and pathways, while adipose distribution is enriched more broadly across physiological systems. In addition, polygenic scores for the adiposity factor predict many adverse health outcomes, while those for body size and composition predict a more limited subset. Finally, we characterize the factors' genetic correlations with obesity-related traits and examine the druggable genome by constructing a bipartite drug-gene network to identify potential therapeutic targets.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Bryan, S. et al. NHSR. National Health and Nutrition Examination Survey 2017–March 2020 Pre-Pandemic Data Files. https://stacks.cdc.gov/view/cdc/106273 (2021).
-
- CDC. Obesity is a common, serious, and costly disease. Cent. Dis. Control Prev.https://www.cdc.gov/obesity/data/adult.html (2022).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

