Complementary cytoskeletal feedback loops control signal transduction excitability and cell polarity
- PMID: 40796751
- PMCID: PMC12344061
- DOI: 10.1038/s41467-025-62799-3
Complementary cytoskeletal feedback loops control signal transduction excitability and cell polarity
Abstract
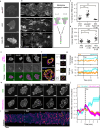
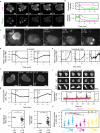
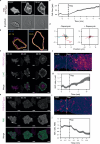
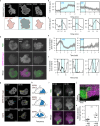
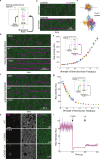
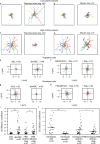
To navigate complex environments, cells integrate chemical and mechanical cues through dynamic feedback between signaling networks and the cytoskeleton. Using synthetic tools to manipulate cytoskeletal components in Dictyostelium and human neutrophils, we uncover feedback mechanisms that regulate Ras/PI3K signaling and control front- and back-states of the cell. Increased branched actin and actin polymerization enhance Ras/PI3K activity. Similarly, decreased myosin II assembly also elevates signaling and chemotactic sensitivity. Conversely, inhibiting branched actin increases cortical actin and blocks Ras/PI3K activation-an effect lessened by decreasing filamentous actin or in myosin II-null cells. Activating RacE to increase actin crosslinking suppresses Ras activity without triggering branched actin nucleators, yet promotes spreading and protrusion. These results informed a computational model incorporating positive cytoskeletal feedback loops, which predicts shifts in polarity and migration with cytoskeletal changes. We propose that such feedback locally tunes signal network excitability, enabling cells to navigate tissues, extracellular matrix, and fluid environments.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Update of
-
Complementary Cytoskeletal Feedback Loops Control Signal Transduction Excitability and Cell Polarity.bioRxiv [Preprint]. 2024 Feb 13:2024.02.13.580131. doi: 10.1101/2024.02.13.580131. bioRxiv. 2024. Update in: Nat Commun. 2025 Aug 12;16(1):7482. doi: 10.1038/s41467-025-62799-3. PMID: 38405988 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 GM066817/GM/NIGMS NIH HHS/United States
- R01 GM149073/GM/NIGMS NIH HHS/United States
- AFOSR MURI FA95501610052/United States Department of Defense | United States Air Force | AFMC | Air Force Research Laboratory (AFRL)
- S10 OD016374/OD/NIH HHS/United States
- DARPA HR0011-16-C-0139/United States Department of Defense | Defense Advanced Research Projects Agency (DARPA)
LinkOut - more resources
Full Text Sources

