Direction and modality of transcription changes caused by TAD boundary disruption in Slc29a3/Unc5b locus depends on tissue-specific epigenetic context
- PMID: 40796890
- PMCID: PMC12341078
- DOI: 10.1186/s13072-025-00618-1
Direction and modality of transcription changes caused by TAD boundary disruption in Slc29a3/Unc5b locus depends on tissue-specific epigenetic context
Abstract
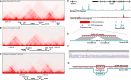
Background: Topologically associating domains (TADs) are believed to play a role in the regulation of gene expression by constraining or guiding interactions between the regulatory elements. While the impact of TAD perturbations is typically studied in developmental genes with highly cell-type-specific expression patterns, this study examines genes with broad expression profiles separated by a strong insulator boundary. We focused on the mouse Slc29a3/Unc5b locus, which encompasses two distinct TADs containing ubiquitously expressed and essential for viability genes. We disrupted the CTCF-boundary between these TADs and analyzed the resulting changes in gene expression.
Results: Deletion of four CTCF binding sites at the TAD boundary altered local chromatin architecture, abolishing pre‑existing loops and creating novel long‑range interactions that spanned the original TAD boundary. Using UMI-assisted targeted RNA-seq we evaluated transcriptional changes of Unc5b, Slc29a3, Psap, Vsir, Cdh23, and Sgpl1 across various organs. We found that TAD boundary disruption led to variable transcriptional responses, where not only the magnitude but also the direction of gene expression changes were tissue-specific. Current hypotheses on genome architecture function, such as enhancer competition and hijacking, as well as genomic deep learning models, only partially explain these transcriptional changes, highlighting the need for further investigation into the mechanisms underlying TAD function and gene regulation.
Conclusions: Disrupting the insulator element between broadly expressed genes resulted in moderate, tissue-dependent transcriptional alterations, rather than uniformly activating or silencing the target genes. These findings show that TAD boundaries contribute to context‑specific regulation even at housekeeping loci and underscore the need for refined models to predict the effects of non‑coding structural variants.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Bystander activation across a TAD boundary supports a cohesin-dependent transcription cluster model for enhancer function.Genes Dev. 2025 Sep 2;39(17-18):1012-1024. doi: 10.1101/gad.352648.125. Genes Dev. 2025. PMID: 40436628 Free PMC article.
-
Linking DNA-packing density distribution and TAD boundary locations.Proc Natl Acad Sci U S A. 2025 Mar 4;122(9):e2418456122. doi: 10.1073/pnas.2418456122. Epub 2025 Feb 25. Proc Natl Acad Sci U S A. 2025. PMID: 39999165 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Maternal and neonatal outcomes of elective induction of labor.Evid Rep Technol Assess (Full Rep). 2009 Mar;(176):1-257. Evid Rep Technol Assess (Full Rep). 2009. PMID: 19408970 Free PMC article.
References
-
- Nuriddinov M, Fishman V. C-InterSecture-a computational tool for interspecies comparison of genome architecture. Bioinforma Oxf Engl. 2019;35(23):4912–21. - PubMed
-
- Fishman V, Battulin N, Nuriddinov M, Maslova A, Zlotina A, Strunov A, et al. 3D organization of chicken genome demonstrates evolutionary conservation of topologically associated domains and highlights unique architecture of erythrocytes’ chromatin. Nucleic Acids Res. 2019;47(2):648–65. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

