Genetic architecture of bone marrow fat fraction implies its involvement in osteoporosis risk
- PMID: 40796918
- PMCID: PMC12344015
- DOI: 10.1038/s41467-025-62826-3
Genetic architecture of bone marrow fat fraction implies its involvement in osteoporosis risk
Abstract
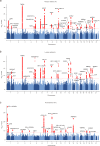
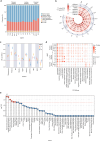
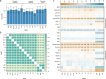
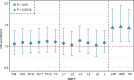
Bone marrow adipose tissue, as a distinct adipose subtype, has been implicated in the pathophysiology of skeletal, metabolic, and hematopoietic disorders. To identify its underlying genetic factors, we utilized a deep learning algorithm capable of quantifying bone marrow fat fraction (BMFF) in the vertebrae and proximal femur using magnetic resonance imaging data of over 38,000 UK Biobank participants. Genome-wide association analyses uncovered 373 significant BMFF-associated variants (P-value < 5 × 10-9), with enrichment in bone remodeling, metabolism, and hematopoiesis pathway. Furthermore, genetic correlation highlighted a significant association between BMFF and skeletal disease. In about 300,000 individuals, polygenic risk scores derived from three proximal femur BMFF were significantly associated with increased osteoporosis risk. Notably, Mendelian randomization analyses revealed a causal link between proximal femur BMFF and osteoporosis. Here, we show critical insights into the genetic determinants of BMFF and offer perspectives on the biological mechanisms driving osteoporosis development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Wang, Y. et al. Bone marrow adipocyte: Origin, biology and relationship with hematological malignancy. Int. J. Lab Hematol.46, 10–19 (2024). - PubMed
-
- Manolagas, S. C. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr. Rev.21, 115–137 (2000). - PubMed
-
- Zhu, L. et al. Marrow adiposity as an indicator for insulin resistance in postmenopausal women with newly diagnosed type 2 diabetes - an investigation by chemical shift-encoded water-fat MRI. Eur. J. Radiol.113, 158–164 (2019). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

