Decoding circRNA translation: challenges and advances in computational method development
- PMID: 40799216
- PMCID: PMC12339343
- DOI: 10.3389/fgene.2025.1654305
Decoding circRNA translation: challenges and advances in computational method development
Abstract
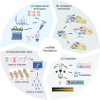
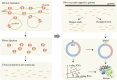
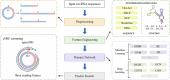
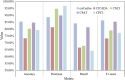
In recent years, numerous studies have demonstrated that circRNAs play crucial biological roles through their capacity to encode functional proteins. Computational methods have become essential for investigating circRNA translation. In this review, we first outline circRNA biogenesis and translation mechanisms to establish the rationale for developing specialized computational strategies. We then summarize experimental techniques and existing databases that support computational method development. Subsequently, we provide a systematic introduction to existing circRNA translation analysis tools and their underlying algorithms, with emphasis on benchmarking the performance of sequence-based methods using a unified dataset. Our benchmarking revealed that: (1) cirCodAn achieved superior predictive accuracy while maintaining user accessibility; (2) the training data selection during method development critically impacts model performance. This review serves as a comprehensive reference for the selection and application of circRNA translation analysis methods and provides foundational guidance for the development and refinement of future computational tools.
Keywords: bioinformatics; circRNA; coding potential; function; translation.
Copyright © 2025 Zhang, Zhou, Zhang, Peng, Meng, Xi and Wei.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Exploring a circulating circRNA and miRNA biomarker panel for early detection of ovarian cancer through multiple omics analysis.Sci Rep. 2025 Jul 16;15(1):25809. doi: 10.1038/s41598-025-11641-3. Sci Rep. 2025. PMID: 40670591 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Survivor, family and professional experiences of psychosocial interventions for sexual abuse and violence: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2022 Oct 4;10(10):CD013648. doi: 10.1002/14651858.CD013648.pub2. Cochrane Database Syst Rev. 2022. PMID: 36194890 Free PMC article.
-
Gender differences in the context of interventions for improving health literacy in migrants: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2024 Dec 12;12(12):CD013302. doi: 10.1002/14651858.CD013302.pub2. Cochrane Database Syst Rev. 2024. PMID: 39665382
-
How lived experiences of illness trajectories, burdens of treatment, and social inequalities shape service user and caregiver participation in health and social care: a theory-informed qualitative evidence synthesis.Health Soc Care Deliv Res. 2025 Jun;13(24):1-120. doi: 10.3310/HGTQ8159. Health Soc Care Deliv Res. 2025. PMID: 40548558
References
-
- Barbosa D. F., Oliveira L. S., Kashiwabara A. Y. (2023). “circTIS: a weighted degree string kernel with support vector machine tool for translation initiation sites prediction in circRNA,” in Advances in Bioinformatics and Computational Biology: 16th Brazilian Symposium on Bioinformatics, BSB 2023, Curitiba, Brazil, June 13–16, 2023, 14–24.
Publication types
LinkOut - more resources
Full Text Sources

