Cargo-selective regulation of clathrin-mediated endocytosis by AMP-activated protein kinase
- PMID: 40799387
- PMCID: PMC12341638
- DOI: 10.1016/j.isci.2025.113131
Cargo-selective regulation of clathrin-mediated endocytosis by AMP-activated protein kinase
Abstract
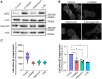
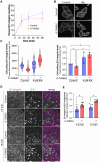
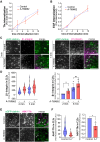
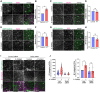
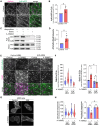
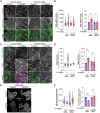
The cell surface abundance of many proteins is controlled by clathrin-mediated endocytosis (CME). CME is driven by the assembly of clathrin and other proteins on the inner leaflet of the plasma membrane into clathrin-coated pits (CCPs). Regulation of CCP dynamics allows for control of the function of specific cell surface proteins, impacting a range of cellular outcomes. AMP-activated protein kinase (AMPK) becomes activated upon metabolic insufficiency and facilitates cellular adaptation to nutrient stress. Here, we examined how AMPK regulates CME and the cell surface membrane traffic of β1-integrin. We find that AMPK controls CCP dynamics and regulates the abundance of the endocytic adaptor protein Dab2 within CCPs in a manner that requires the GTPase Arf6, thus selectively promoting the CCP recruitment and internalization of β1-integrin. This study reveals a signaling pathway for cargo-selective metabolic regulation of CME by AMPK that impacts the function of cell surface proteins such as integrins.
Keywords: Biochemistry; Cell biology.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources

