Estimating protein complex model accuracy using graph transformers and pairwise similarity graphs
- PMID: 40799498
- PMCID: PMC12342149
- DOI: 10.1093/bioadv/vbaf180
Estimating protein complex model accuracy using graph transformers and pairwise similarity graphs
Abstract
Motivation: Estimation of protein complex structure accuracy is essential for effective structural model selection in structural biology applications such as protein function analysis and drug design. Despite the success of structure prediction methods such as AlphaFold2 and AlphaFold3, selecting top-quality structural models from large model pools remains challenging.
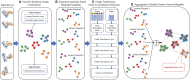
Results: We present GATE, a novel method that uses graph transformers on pairwise model similarity graphs to predict the quality (accuracy) of complex structural models. By integrating single-model and multimodel quality features, GATE captures intrinsic model characteristics and intermodel geometric similarities to make robust predictions. On the dataset of the 15th Critical Assessment of Protein Structure Prediction (CASP15), GATE achieved the highest Pearson's correlation (0.748) and the lowest ranking loss (0.1191) compared with existing methods. In the blind CASP16 experiment, GATE ranked fifth based on the sum of z-scores, with a Pearson's correlation of 0.7076 (first), a Spearman's correlation of 0.4514 (fourth), a ranking loss of 0.1221 (third), and an area under the curve score of 0.6680 (third) on per-target TM-score-based metrics. Additionally, GATE also performed consistently on large in-house datasets generated by extensive AlphaFold-based sampling with MULTICOM4, confirming its robustness and practical applicability in real-world model selection scenarios.
Availability and implementation: GATE is available at https://github.com/BioinfoMachineLearning/GATE.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
Update of
-
Estimating Protein Complex Model Accuracy Using Graph Transformers and Pairwise Similarity Graphs.bioRxiv [Preprint]. 2025 Feb 23:2025.02.04.636562. doi: 10.1101/2025.02.04.636562. bioRxiv. 2025. Update in: Bioinform Adv. 2025 Jul 29;5(1):vbaf180. doi: 10.1093/bioadv/vbaf180. PMID: 39975041 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources

