This is a preprint.
Proteomic signatures of the APOE ε 4 and APOE ε 2 genetic variants and Alzheimer's disease
- PMID: 40799961
- PMCID: PMC12340876
- DOI: 10.1101/2025.08.04.25332945
Proteomic signatures of the APOE ε 4 and APOE ε 2 genetic variants and Alzheimer's disease
Abstract
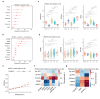
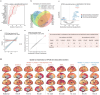
The ε4 and ε2 alleles of the Apolipoprotein E (APOE) gene confer opposite genetic risks for Alzheimer's disease (AD), but their underlying molecular mechanisms remain poorly characterized in humans. To resolve this, we systematically profiled APOE-associated proteomic alterations across five cohorts-including the Global Neurodegeneration Proteomics Consortium (GNPC), BioFINDER-2, the Alzheimer's Disease Neuroimaging Initiative (ADNI), the Parkinson's Progression Markers Initiative (PPMI), and UK Biobank (UKB)-using SomaLogic and OLINK platforms in plasma and cerebrospinal fluid (CSF) from over 10,000 individuals. Using GNPC (plasma SomaLogic, N=4,045), we mapped a comprehensive APOE-protein network and applied mediation modeling to classify genotype-related signals as upstream mediators, downstream consequences, or APOE-specific changes. We then leveraged CSF beta-amyloid (Aβ) biomarker data from BioFINDER-2 (plasma SomaLogic, N=1,421) to improve temporal resolution and isolate early, Aβ-independent proteomic programs. In the Aβ- individuals, APOE4 was linked to cell cycle and chromatin remodeling, while APOE2 was associated with mitochondrial regulation and DNA repair. Mediation analyses nominated proteins such as S100A13, TBCA, SPC25 for APOE4, and APOB, SNAP23 for APOE2 as candidate upstream effectors, supported by CSF validation (ADNI, SomaLogic, N=666), brain transcriptomic co-expression, and AD GWAS colocalization. Longitudinal CSF data from PPMI confirmed the temporal stability of several APOE-associated proteins. Cross-platform comparisons (UKB plasma OLINK, N=4,820, and BF2 CSF OLINK, N=1,475) revealed matrix- and assay-specific heterogeneity, underscoring challenges in reproducibility. Together, our results delineate allele-specific, temporally structured proteomic signatures that precede AD pathology, offering insight into APOE-driven molecular pathways and potential therapeutic targets for early intervention.
Conflict of interest statement
Ethics disclosures Conflicts of interest NMC has received speaker/consultancy fees from Biogen, Eli Lilly, Owkin and Merck. OH is an employee of Lund University and Eli Lilly. SP has acquired research support (for the institution) from Avid and ki elements through ADDF. In the past 2 years, he has received consultancy/speaker fees from Bioartic, Biogen, Eisai, Eli Lilly, Novo Nordisk, and Roche. BM and AV are employees of Abbvie. JWV has received consultancy fees from Manifest Technologies.
Figures




References
-
- Corder E. H. et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923 (1993). - PubMed
-
- Mattsson N. et al. Prevalence of the apolipoprotein E ε4 allele in amyloid β positive subjects across the spectrum of Alzheimer’s disease. Alzheimers. Dement. 14, 913–924 (2018). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
