Overcoming challenges in metagenomic AMR surveillance with nanopore sequencing: a case study on fluoroquinolone resistance
- PMID: 40800117
- PMCID: PMC12342191
- DOI: 10.3389/fmicb.2025.1614301
Overcoming challenges in metagenomic AMR surveillance with nanopore sequencing: a case study on fluoroquinolone resistance
Abstract
Introduction: Antimicrobial resistance is an alarming public health problem, and comprehensive surveillance across environments is required to reduce its impact. Phenotypic testing and whole-genome sequencing of isolates are efficient, but culture-free approaches like metagenomic sequencing potentially allow for broader investigation of resistance gene occurrence, evolution and spread. However, technical challenges such as difficulties in associating antimicrobial resistance genes with their bacterial hosts and the collapse of strain-level variation during metagenome assembly, hinder its implementation.
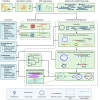
Methods: To illustrate how these challenges can be overcome, we applied Oxford Nanopore Technologies long-read metagenomic sequencing and novel bioinformatic methods to a case study focused on fluoroquinolone resistance in chicken fecal samples.
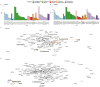
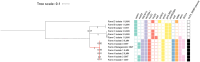
Results: We demonstrate plasmid-host linking based on detecting common DNA methylation signatures. Additionally, we use new bioinformatic approaches for strain haplotyping, enabling phylogenomic comparison and uncovering fluoroquinolone resistance determining point mutations in metagenomic datasets.
Discussion: We leverage long-read sequencing, including DNA methylation profiling and strain-level haplotyping, to identify antimicrobial resistance gene hosts, link plasmids to their bacterial carriers, and detect resistance-associated point mutations. Although some limitations remain, our work demonstrates how these improvements in metagenomic sequencing can enhance antimicrobial resistance surveillance.
Keywords: DNA methylation; antimicrobial resistance (AMR); metagenomic sequencing; nanopore sequencing; plasmid host prediction; strain-resolved metagenomics.
Copyright © 2025 Bloemen, Gand, Ringenier, Bogaerts, Vanneste, Marchal, Roosens, Dewulf, Boyen and De Keersmaecker.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- BELMAP . (2024). BELMAP 2024: One Health Report of Antimicrobial Consumption and Resistance in Belgium. Brussels: BELMAP . 10.25608/v1x6-1e19 - DOI
LinkOut - more resources
Full Text Sources

