Evidence for a compensatory relationship between left- and right-lateralized brain networks
- PMID: 40800905
- PMCID: PMC12319987
- DOI: 10.1162/imag_a_00437
Evidence for a compensatory relationship between left- and right-lateralized brain networks
Abstract
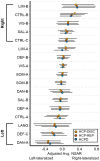
The two hemispheres of the human brain are functionally asymmetric. At the network level, the language network exhibits left-hemisphere lateralization. While this asymmetry is widely replicated, the extent to which other functional networks demonstrate lateralization remains a subject of investigation. Additionally, it is unknown how the lateralization of one functional network may affect the lateralization of other networks within individuals. We quantified lateralization for each of 17 networks by computing the relative surface area on the left and right cerebral hemispheres. After examining the ecological, convergent, and external validity and test-retest reliability of this surface area-based measure of lateralization, we addressed two hypotheses across multiple datasets (Human Connectome Project = 553, Human Connectome Project-Development = 343, Natural Scenes Dataset = 8). First, we hypothesized that networks associated with language, visuospatial attention, and executive control would show the greatest lateralization. Second, we hypothesized that relationships between lateralized networks would follow a dependent relationship such that greater left lateralization of a network would be associated with greater right lateralization of a different network within individuals, and that this pattern would be systematic across individuals. A language network was among the three networks identified as being significantly left lateralized, and attention and executive control networks were among the five networks identified as being significantly right lateralized. Next, correlation matrices, an exploratory factor analysis, and confirmatory factor analyses were used to test the second hypothesis and examine the organization of lateralized networks. We found general support for a dependent relationship between highly left- and right-lateralized networks, meaning that across subjects, greater left lateralization of a given network (such as a language network) was linked to greater right lateralization of another network (such as a ventral attention/salience network) and vice versa. These results further our understanding of brain organization at the macro-scale network level in individuals, carrying specific relevance for neurodevelopmental conditions characterized by disruptions in lateralization such as autism and schizophrenia.
Keywords: asymmetry; attention; brain networks; fMRI; language; lateralization.
© 2025 The Authors. Published under a Creative Commons Attribution 4.0 International (CC BY 4.0) license.
Conflict of interest statement
The authors have no known conflict of interest to disclose.
Figures




Update of
-
Evidence for a Compensatory Relationship between Left- and Right-Lateralized Brain Networks.bioRxiv [Preprint]. 2023 Dec 9:2023.12.08.570817. doi: 10.1101/2023.12.08.570817. bioRxiv. 2023. Update in: Imaging Neurosci (Camb). 2025 Jan 29;3:imag_a_00437. doi: 10.1162/imag_a_00437. PMID: 38106130 Free PMC article. Updated. Preprint.
References
-
- Allen , E. J. , St-Yves , G. , Wu , Y. , Breedlove , J. L. , Prince , J. S. , Dowdle , L. T. , Nau , M. , Caron , B. , Pestilli , F. , Charest , I. , Hutchinson , J. B. , Naselaris , T. , & Kay , K. ( 2022. ). A massive 7T fMRI dataset to bridge cognitive neuroscience and artificial intelligence . Nature Neuroscience , 25 ( 1 ), Article 1. 10.1038/s41593-021-00962-x - DOI - PubMed
-
- Andrew , R. J. , Mench , J. , & Rainey , C. ( 1982. ). Right-left asymmetry of response to visual stimuli in the domestic chick . In Ingle D. J. , Goodale M. A. , & Mansfield R. J. W. , (Eds.), Analysis of Visual Behaviour , (pp. 197 – 209 ). MIT Press; . ISBN: 9780262090223.
-
- Anticevic , A. , Dierker , D. L. , Gillespie , S. K. , Repovs , G. , Csernansky , J. G. , Van Essen , D. C. , & Barch , D. M. ( 2008. ). Comparing surface-based and volume-based analyses of functional neuroimaging data in patients with schizophrenia . NeuroImage , 41 ( 3 ), 835 – 848 . 10.1016/j.neuroimage.2008.02.052 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
