Museum Genomics Reveals Temporal Genetic Stasis and Global Genetic Diversity in Arabidopsis thaliana
- PMID: 40801356
- PMCID: PMC12530293
- DOI: 10.1111/mec.70081
Museum Genomics Reveals Temporal Genetic Stasis and Global Genetic Diversity in Arabidopsis thaliana
Abstract
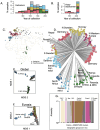
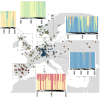
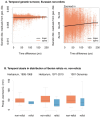
Global patterns of population genetic variation through time offer a window into evolutionary processes that maintain diversity. Over time, lineages may expand or contract their distribution, causing turnover in population genetic composition. At individual loci, migration, drift and selection (among other processes) may affect allele frequencies. Museum specimens of widely distributed species offer a unique window into the genetics of understudied populations and changes over time. Here, we sequenced genomes of 130 herbarium specimens and 91 new field collections of Arabidopsis thaliana and combined these with published genomes. We sought a broader view of genomic diversity across the species and to test if population genomic composition is changing through time. We documented extensive and previously uncharacterised diversity in a range of populations in Africa, populations that are under threat from anthropogenic climate change. Through time, we did not find dramatic changes in genomic composition of populations. Instead, we found a pattern of genetic change every 100 years of the same magnitude seen when comparing Eurasian populations that are 185 km apart, potentially due to a combination of drift and changing selection. We found only mixed signals of polygenic adaptation at phenology and physiology QTL. We did find that genes conserved across eudicots show altered levels of directional allele frequency change, potentially due to variable purifying and background selection. Our study highlights how museum specimens can reveal new dimensions of population diversity and show how wild populations are evolving in recent history.
Keywords: allele frequency change; genomic diversity; museum specimens; population structure; time series genomics.
© 2025 The Author(s). Molecular Ecology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Update of
-
Museum genomics reveals temporal genetic stasis and global genetic diversity in Arabidopsis thaliana.bioRxiv [Preprint]. 2025 Feb 7:2025.02.06.636844. doi: 10.1101/2025.02.06.636844. bioRxiv. 2025. Update in: Mol Ecol. 2025 Oct;34(20):e70081. doi: 10.1111/mec.70081. PMID: 39975324 Free PMC article. Updated. Preprint.
References
-
- Ågren, J. , and Schemske D. W.. 2012. “Reciprocal Transplants Demonstrate Strong Adaptive Differentiation of the Model Organism <styled-content style="fixed-case"> Arabidopsis thaliana </styled-content> in Its Native Range.” New Phytologist 194, no. 4: 1112–1122. 10.1111/j.1469-8137.2012.04112.x. - DOI - PubMed
-
- Akbari, A. , Barton A. R., Gazal S., et al. 2024. “Pervasive Findings of Directional Selection Realize the Promise of Ancient DNA to Elucidate Human Adaptation.” Preprint, bioRxiv, September 15. 10.1101/2024.09.14.613021. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

