m6A-Modified circRAPGEF1 Interaction with IGF2BP3 Promotes Hepatocellular Carcinoma Progression via Reprogramming Aspartate Metabolism
- PMID: 40801445
- PMCID: PMC12591180
- DOI: 10.1002/advs.202503851
m6A-Modified circRAPGEF1 Interaction with IGF2BP3 Promotes Hepatocellular Carcinoma Progression via Reprogramming Aspartate Metabolism
Abstract
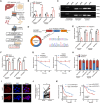
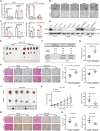
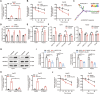
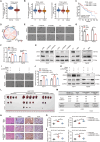
Hepatocellular carcinoma (HCC) progression and therapy sensitivity are critically fueled by liver cancer stem cells (LCSCs), yet the regulatory mechanisms of circular RNAs (circRNAs) on LCSCs remain elusive. Here, through circRNA microarray analysis of LCSCs and non-stem HCC cells, circRAPGEF1 is identified as a LCSC-enriched circRNA upregulated in HCC tissues and predictive of poor patient survival. Functionally, circRAPGEF1 promoted the stemness properties, proliferation, and tumorigenicity of HCC cells. Mechanistically, the METTL3-mediated N6-methyladenosine (m6A) modification of circRAPGEF1 facilitated KH domain-dependent binding of IGF2BP3 to its UGGAC motif, which conferring stability to circRAPGEF1 while competitively disrupting the IGF2BP3/ASS1 mRNA interaction. This process led to the degradation of ASS1 mRNA, triggering aspartate accumulation and activation of the S6K/CAD signaling pathway. Crucially, circRAPGEF1 overexpression reduced the sorafenib sensitivity, whereas targeting circRAPGEF1 using nanoparticles-mediated systematic siRNAs delivery effectively sensitized HCC cells to sorafenib. Collectively, these findings unveil a METTL3/circRAPGEF1/IGF2BP3/ASS1 regulatory axis that drives aspartate metabolic reprogramming to fuel HCC stemness properties, positioning circRAPGEF1 as a dual prognostic biomarker and therapeutic target to enhance sorafenib efficacy in HCC.
Keywords: aspartate metabolism; circular RNAs; hepatocellular carcinoma; liver cancer stem cells; m6A modification.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bray F., Laversanne M., Sung H., Ferlay J., Siegel R. L., Soerjomataram I., Jemal A., Ca‐Cancer J. Clin. 2024, 74, 229. - PubMed
-
- Singal A. G., Kanwal F., Llovet J. M., Nat. Rev. Clin. Oncol. 2023, 20, 864. - PubMed
-
- Lee T. K., Guan X., Ma S., Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 26. - PubMed
-
- Ma S., Chan K.‐W., Hu L., Lee T. K.‐W., Wo J. Y.‐H., Ng I. O.‐L., Zheng B.‐J., Guan X.‐Y., Gastroenterology 2007, 132, 2542. - PubMed
-
- Liu C., Chen L., Cell 2022, 185, 2016. - PubMed
MeSH terms
Substances
Grants and funding
- 81902413/National Natural Science Foundation of China
- 82073045/National Natural Science Foundation of China
- 202201020311/Science and Technology Program of Guangzhou
- 2024A1515010579/Guangdong Basic and Applied Basic Research Foundation
- 2023A1515010745/Guangdong Basic and Applied Basic Research Foundation
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
