Generation of Individualized, Standardized, and Electrically Synchronized Human Midbrain Organoids
- PMID: 40801645
- PMCID: PMC12346388
- DOI: 10.3390/cells14151211
Generation of Individualized, Standardized, and Electrically Synchronized Human Midbrain Organoids
Abstract
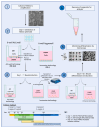
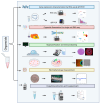
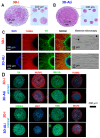
Organoids allow to model healthy and diseased human tissues. and have applications in developmental biology, drug discovery, and cell therapy. Traditionally cultured in immersion/suspension, organoids face issues like lack of standardization, fusion, hypoxia-induced necrosis, continuous agitation, and high media volume requirements. To address these issues, we developed an air-liquid interface (ALi) technology for culturing organoids, termed AirLiwell. It uses non-adhesive microwells for generating and maintaining individualized organoids on an air-liquid interface. This method ensures high standardization, prevents organoid fusion, eliminates the need for agitation, simplifies media changes, reduces media volume, and is compatible with Good Manufacturing Practices. We compared the ALi method to standard immersion culture for midbrain organoids, detailing the process from human pluripotent stem cell (hPSC) culture to organoid maturation and analysis. Air-liquid interface organoids (3D-ALi) showed optimized size and shape standardization. RNA sequencing and immunostaining confirmed neural/dopaminergic specification. Single-cell RNA sequencing revealed that immersion organoids (3D-i) contained 16% fibroblast-like, 23% myeloid-like, and 61% neural cells (49% neurons), whereas 3D-ALi organoids comprised 99% neural cells (86% neurons). Functionally, 3D-ALi organoids showed a striking electrophysiological synchronization, unlike the heterogeneous activity of 3D-i organoids. This standardized organoid platform improves reproducibility and scalability, demonstrated here with midbrain organoids. The use of midbrain organoids is particularly relevant for neuroscience and neurodegenerative diseases, such as Parkinson's disease, due to their high incidence, opening new perspectives in disease modeling and cell therapy. In addition to hPSC-derived organoids, the method's versatility extends to cancer organoids and 3D cultures from primary human cells.
Keywords: 3D cell culture; AirLiwell; Parkinson’s disease; air–liquid interface; cell therapy; electrical recordings; neurospheres; organoids; pluripotent stem cells; spheroids.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
A Hybrid 2D-to-3D in vitro Differentiation Platform Improves Outcomes of Cerebral Cortical Organoid Generation in hiPSCs.Curr Protoc. 2024 Oct;4(10):e70022. doi: 10.1002/cpz1.70022. Curr Protoc. 2024. PMID: 39400999
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Optimized scaffold-free human 3D adipose tissue organoid culture for obesity and disease modeling.SLAS Discov. 2025 Mar;31:100218. doi: 10.1016/j.slasd.2025.100218. Epub 2025 Jan 25. SLAS Discov. 2025. PMID: 39870353
-
Advances, challenges, and opportunities of human midbrain organoids for modelling of the dopaminergic system.EMBO J. 2025 Aug;44(15):4181-4195. doi: 10.1038/s44318-025-00494-1. Epub 2025 Jul 2. EMBO J. 2025. PMID: 40604323 Free PMC article. Review.
-
Potential Use of Organoids in Regenerative Medicine.Tissue Eng Regen Med. 2024 Dec;21(8):1125-1139. doi: 10.1007/s13770-024-00672-y. Epub 2024 Oct 16. Tissue Eng Regen Med. 2024. PMID: 39412646 Review.
References
-
- Jo J., Xiao Y., Sun A.X., Cukuroglu E., Tran H.D., Goke J., Tan Z.Y., Saw T.Y., Tan C.P., Lokman H., et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

