High-risk clonal groups of avian pathogenic Escherichia coli (APEC) demonstrate heterogeneous phenotypic characteristics in vitro and in vivo
- PMID: 40801816
- PMCID: PMC12351747
- DOI: 10.1080/21505594.2025.2546682
High-risk clonal groups of avian pathogenic Escherichia coli (APEC) demonstrate heterogeneous phenotypic characteristics in vitro and in vivo
Abstract
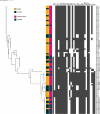
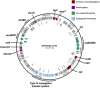
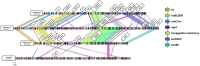
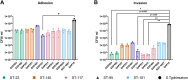
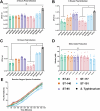
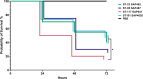
Avian Pathogenic Escherichia coli (APEC), a major bacterial pathogen of poultry, comprises a diverse range of high-risk clonal groups. However how these lineages interact with avian host cells remains poorly characterized. This study examined the ability of key APEC clonal groups to adhere to, invade, and survive within avian host cells, alongside assessing their virulence in the Galleria mellonella infection model. Genomic analysis of APEC from a UK turkey colibacillosis outbreak identified ST-101 as the dominant clonal group, carrying numerous virulence factors. ST-101 was compared to other high-risk APEC clonal groups (ST-23, ST-140, ST-95, ST-117). Utilizing in vitro cell culture models, APEC isolates displayed comparable adhesion to 8E11 chicken epithelial gut and HD11 chicken macrophage cell lines. APEC ST-95, ST-101, and ST-140 demonstrated increased invasion of 8E11 cells, and intracellular survival within HD11 macrophages, relative to ST-23 and ST-117, suggesting pronounced phenotypic differences between clonal groups. However, in HD11 cell assays, no difference in magnitude of elicited immune response was observed between lineages, indicating differing intracellular survival was not a result of immune response modulation. In vivo virulence in the Galleria mellonella infection model was also observed to differ between APEC genotypes, with ST-117 inducing the highest mortality, despite the comparatively lower epithelial invasion and intramacrophage survival for other lineages. Collectively, this suggests that diverse APEC genotypes have distinct phenotypic profiles in vitro and in vivo. These results highlight the need for intervention strategies that can simultaneously target a broad range of pathogenic lineages.
Keywords: APEC; Escherichia coli; Galleria mellonella; avian pathogenic; cell culture; sequence types.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
Similar articles
-
Genomic epidemiology of avian pathogenic Escherichia coli in two broiler colibacillosis outbreaks in Finland.Vet Microbiol. 2025 Sep;308:110617. doi: 10.1016/j.vetmic.2025.110617. Epub 2025 Jun 23. Vet Microbiol. 2025. PMID: 40633272
-
Prevalence of specific serogroups, antibiotic resistance and virulence factors of avian pathogenic Escherichia coli (APEC) isolated from clinical cases: A systematic review and meta-analysis.Microb Pathog. 2024 Sep;194:106843. doi: 10.1016/j.micpath.2024.106843. Epub 2024 Aug 6. Microb Pathog. 2024. PMID: 39117015
-
Palygorskite improves growth performance and prevents liver damage in avian pathogenic Escherichia coli-challenged broiler chickens at an early age.J Anim Sci. 2024 Jan 3;102:skae302. doi: 10.1093/jas/skae302. J Anim Sci. 2024. PMID: 39373204
-
Phenotypic and genotypic characterization of biofilm-producing avian pathogenic Escherichia coli (APEC) isolates from Algerian poultry: associations between antimicrobial resistance and virulence genes.Vet Res Commun. 2025 Jun 21;49(4):232. doi: 10.1007/s11259-025-10801-0. Vet Res Commun. 2025. PMID: 40542899
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
References
LinkOut - more resources
Full Text Sources
