The emergence of genetic variants linked to brain and cognitive traits in human evolution
- PMID: 40801890
- PMCID: PMC12345208
- DOI: 10.1093/cercor/bhaf127
The emergence of genetic variants linked to brain and cognitive traits in human evolution
Abstract
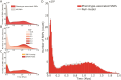
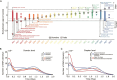
Human evolution involved major anatomical transformations, including a rapid increase in brain volume over the last 2 million years. Examination of fossil records provides insight into these physical changes but offers limited information on the evolution of brain function and cognition. A complementary approach integrates genome dating from the Human Genome Dating Project with genome-wide association studies to trace the emergence of genetic variants linked to human traits over 5 million years. We find that genetic variants underlying cortical morphology (~300,000 years, P = 4 × 10-28), fluid intelligence (~500,000 years, P = 1.4 × 10-4), and psychiatric disorders (~475,000 years, P = 5.9 × 10-33) emerged relatively recently in hominin evolution. Among psychiatric phenotypes, variants associated with depression (~24,000 years, P = 1.6 × 10-4) and alcoholism-related traits (~40,000 years, P = 5.2 × 10-12) are the youngest. Genes with recent evolutionary modifications are involved in intelligence (P = 1.7 × 10-6) and cortical area (P = 3.5 × 10-4) and exhibit elevated expression in language-related areas (P = 7.1 × 10-4), a hallmark of human cognition. Our findings suggest that recently evolved genetic variants shaped the human brain, cognition, and psychiatric traits.
Keywords: Paleogenomics; brain; evolution; genetics; neuropsychiatry.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
MPvdH is involved as a data consultant for ROCHE and acts as an editor for Wiley Human Brain Mapping. All other authors report no biomedical financial interests or potential conflicts of interest.
Figures

Similar articles
-
Identification of genes associated with dissociation of cognitive performance and neuropathological burden: Multistep analysis of genetic, epigenetic, and transcriptional data.PLoS Med. 2017 Apr 25;14(4):e1002287. doi: 10.1371/journal.pmed.1002287. eCollection 2017 Apr. PLoS Med. 2017. PMID: 28441426 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Structural variants linked to Alzheimer's disease and other common age-related clinical and neuropathologic traits.Genome Med. 2025 Mar 4;17(1):20. doi: 10.1186/s13073-025-01444-6. Genome Med. 2025. PMID: 40038788 Free PMC article.
-
A Systematic Review of the Human Accelerated Regions in Schizophrenia and Related Disorders: Where the Evolutionary and Neurodevelopmental Hypotheses Converge.Int J Mol Sci. 2023 Feb 10;24(4):3597. doi: 10.3390/ijms24043597. Int J Mol Sci. 2023. PMID: 36835010 Free PMC article.
-
Insights into brain evolution through the genotype-phenotype connection.Prog Brain Res. 2023;275:73-92. doi: 10.1016/bs.pbr.2022.12.013. Epub 2023 Feb 3. Prog Brain Res. 2023. PMID: 36841571 Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

