OsROXY2 Regulates Stamen Number Through Interaction with OsbZIP47 in Rice
- PMID: 40801995
- PMCID: PMC12350981
- DOI: 10.1186/s12284-025-00833-0
OsROXY2 Regulates Stamen Number Through Interaction with OsbZIP47 in Rice
Abstract
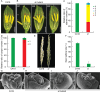
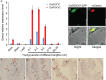
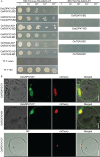
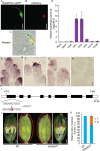
Precise regulation of floral primordia initiation is essential for normal flower development. However, the mechanisms regulating floral primordia initiation (PI) are complex and poorly understood. Herein, we identified a natural mutant in rice, stamen less (sl), which develops florets with reduced stamen number and no carpel due to defects in stamen and carpel PI. STAMENLESS (SL) encodes the CC-type glutaredoxin OsROXY2 and is involved in the regulation of stamen PI. OsROXY1, the closest homolog of OsROXY2, showed no function in stamen PI regulation. The osroxy1 single mutant showed normal reproductive development, while the floret phenotypes of osroxy1/2 double mutant were comparable to those of osroxy2 mutant. The TGA transcription factor OsbZIP47 showed a strong interaction with OsROXY2, and the two genes exhibited overlapping subcellular localizations and expression patterns during flower development. The number of stamens in the osbzip47 mutant was increased to seven (around 35%), indicating that OsbZIP47 is a negative regulator of stamen PI, in contrast to OsROXY2. Taken together, our results reveal that OsROXY2 regulates stamen number via interaction with OsbZIP47, indicating GRX-TGA-mediated floral organ number regulation mechanism is conserved in monocots and eudicots.
Keywords: OsROXY2; OsbZIP47; Floret development; Rice; Stamen number.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing Interests: The authors declare no competing interests.
Figures


Similar articles
-
Androecium homologies in eight-staminate maples: a developmental study.J Plant Res. 2025 Jul;138(4):603-624. doi: 10.1007/s10265-025-01641-9. Epub 2025 Apr 25. J Plant Res. 2025. PMID: 40281254
-
Primary Auxin Response Genes GH3s and DAO1 Modulate Stamen Elongation in Arabidopsis thaliana and Solanum lycopersicum.Physiol Plant. 2025 May-Jun;177(3):e70340. doi: 10.1111/ppl.70340. Physiol Plant. 2025. PMID: 40534123 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Pharmacological interventions for those who have sexually offended or are at risk of offending.Cochrane Database Syst Rev. 2015 Feb 18;2015(2):CD007989. doi: 10.1002/14651858.CD007989.pub2. Cochrane Database Syst Rev. 2015. PMID: 25692326 Free PMC article.
-
Topical antiseptics for chronic suppurative otitis media.Cochrane Database Syst Rev. 2025 Jun 9;6(6):CD013055. doi: 10.1002/14651858.CD013055.pub3. Cochrane Database Syst Rev. 2025. PMID: 40484403
References
-
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr W (2005) Thick Tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132:1235–1245 - PubMed
-
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7:417–427 - PubMed
Grants and funding
- 32260496/National Natural Science Foundation of China
- 32160456/National Natural Science Foundation of China
- Qiankehejichu-ZK[2023]yiban177/Guizhou Provincial Basic Research Program (Natural Science)
- Qianjiaoji (2023) 007/Key Laboratory of Functional Agriculture of Guizhou Provincial Higher Education Institutions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

