KRAS4B is required for placental development
- PMID: 40802112
- PMCID: PMC12350891
- DOI: 10.1007/s00018-025-05846-y
KRAS4B is required for placental development
Abstract
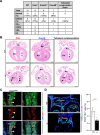
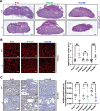
Beyond its well-established role in cancer, KRAS is also crucial for embryogenesis, as its absence leads to embryonic lethality. However, the precise mechanisms underlying the developmental functions of KRAS, as well as the respective roles of its two splicing isoforms, KRAS4A and KRAS4B, remain incompletely characterized. To address these issues, we generated Kras4A knock-out (Kras4A-/-) and Kras4B-/- mouse models using CRISPR/Cas9 technology, and compared their phenotypes to those of a Kras-/- model, in which both isoforms are simultaneously inactivated. We observed that Kras-/- and Kras4B-/- embryos show a lethality that starts around E13.5, while Kras4A-/- embryos develop normally, with no detectable abnormalities. In contrast, Kras-/- embryos displayed a dual phenotype affecting both the heart and placenta, whereas Kras4B-/- embryos exhibited only the placental phenotype. The cardiac phenotype was complex, combining ventricular non-compaction, ventricular septal defects, double outlet right ventricle, and overriding aorta, likely resulting from impaired cardiac precursor proliferation. The placental phenotype was characterized by reduced placental size, and a marked decrease in glycogen trophoblast cells, correlating with hypoglycemia and hypoxia in Kras-/- and Kras4B-/- embryos. Thus, our findings confirm the predominant role of KRAS4B in KRAS-mediated developmental functions, but also suggest hidden functions of KRAS4A. Importantly, this study is the first to identify KRAS as a key regulator of a specific cell differentiation process and to characterize the biological defects caused by its loss.
Keywords: Development; Heart; KRAS4A; KRAS4B; Placenta.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
The differential interactomes of the KRAS splice variants identify BIRC6 as a ubiquitin ligase for KRAS4A.Cell Rep. 2025 Jan 28;44(1):115087. doi: 10.1016/j.celrep.2024.115087. Epub 2024 Dec 19. Cell Rep. 2025. PMID: 39705142 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Can we improve time to patency with vasoepididymostomy with an innovative epididymal occlusion stitch?Int Braz J Urol. 2024 Jul-Aug;50(4):504-506. doi: 10.1590/S1677-5538.IBJU.2024.0222. Int Braz J Urol. 2024. PMID: 38743068 Free PMC article.
References
-
- Assi M, Achouri Y, Loriot A, Dauguet N, Dahou H, Baldan J, Libert M, Fain JS, Guerra C, Bouwens L, Barbacid M, Lemaigre FP, Jacquemin P (2021) Dynamic regulation of expression of KRAS and its effectors determines the ability to initiate tumorigenesis in pancreatic acinar cells. Cancer Res 81:2679–2689 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

