Proviruses in CD4+ T cells reactive to autologous antigens contribute to nonsuppressible HIV-1 viremia
- PMID: 40802737
- PMCID: PMC12363666
- DOI: 10.1126/scitranslmed.adu4643
Proviruses in CD4+ T cells reactive to autologous antigens contribute to nonsuppressible HIV-1 viremia
Abstract
Antiretroviral therapy (ART) halts human immunodeficiency virus-1 (HIV-1) replication, reducing plasma virus concentrations to below the limit of detection, but it is not curative because of a reservoir of latently infected CD4+ T cells. In some people living with HIV-1 (PLWH), plasma HIV-1 RNA becomes persistently detectable despite optimal ART. This nonsuppressible viremia (NSV) is characterized by identical, nonevolving HIV-1 RNA variants expressed from infected CD4+ T cell clones. The mechanisms driving persistent virus production from a specific population of infected cells are poorly understood. We hypothesized that proviruses in cells responding to chronic immunologic stimuli, including self-associated antigens, may drive viral gene expression and NSV. Here, we demonstrate that stimulation of CD4+ T cells with autologous cell lysates induced virus production in a major histocompatibility complex class II-dependent manner. In seven of eight participants with NSV, we recovered viral RNA released ex vivo in response to autologous cell lysates that matched plasma virus. This process involved both defective and replication-competent proviruses residing in conventional CD4+ T cells and was also observed in PLWH with undetectable viremia. These findings suggest that recognition of self-associated antigens is a potentially important cause of HIV-1 reservoir expression, which can contribute to persistent systemic inflammation and rebound upon ART interruption.
Conflict of interest statement
Figures

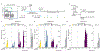




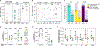
References
-
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M, Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373, 123–126 (1995). - PubMed
-
- Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD, Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387, 188–191 (1997). - PubMed
-
- Chun TW, Davey RT Jr., Engel D, Lane HC, Fauci AS, Re-emergence of HIV after stopping therapy. Nature 401, 874–875 (1999). - PubMed
-
- Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF, In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med 1, 1284–1290 (1995). - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF, Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

