Elusive tropical forest canopy diversity revealed through environmental DNA contained in rainwater
- PMID: 40802774
- PMCID: PMC12346338
- DOI: 10.1126/sciadv.adx4909
Elusive tropical forest canopy diversity revealed through environmental DNA contained in rainwater
Abstract
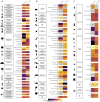
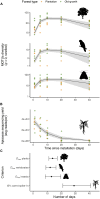
Exploring the biodiversity hidden in tropical rainforests canopies represents a major frontier in biodiversity research yet remains challenging. Environmental DNA (eDNA) can revolutionize this field as it did already in various ecosystems. Here, we test the hypothesis that eDNA contained in canopy throughfall could be used to monitor this elusive diversity and detect anthropogenic disturbance. Using custom-made, low-cost rain collectors, we sampled rainwash eDNA in a mature Amazonian forest and a nearby tree plantation. We successfully detected eDNA from tropical woody and epiphyte plants, vertebrates (mammals, birds, and amphibians), and insects (e.g., mosquitoes, ants, and beetles). The taxonomic composition and diversity reflected disturbance, with significantly lower diversity in the plantation. Crucially, rainwash eDNA integrated biodiversity over a 10-day period in passive collectors and provided a local signature. This approach has thus potential for establishing a cost-effective monitoring system for tropical moist forest canopies, applicable in impact assessments and sustainable management.
Figures


References
-
- FAO, The State of the World’s Biodiversity for Food and Agriculture (FAO Commission on Genetic Resources for Food and Agriculture Assessments, 2019).
-
- IPBES, The IPBES Regional Assessment Report on Biodiversity and Ecosystem Services for the Americas (Secretariat of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services, 2018).
-
- Lapola D. M., Pinho P., Barlow J., Aragão L. E. O. C., Berenguer E., Carmenta R., Liddy H. M., Seixas H., Silva C. V. J., Silva-Junior C. H. L., Alencar A. A. C., Anderson L. O., Armenteras D., Brovkin V., Calders K., Chambers J., Chini L., Costa M. H., Faria B. L., Fearnside P. M., Ferreira J., Gatti L., Gutierrez-Velez V. H., Han Z., Hibbard K., Koven C., Lawrence P., Pongratz J., Portela B. T. T., Rounsevell M., Ruane A. C., Schaldach R., da Silva S. S., von Randow C., Walker W. S., The drivers and impacts of Amazon forest degradation. Science 379, eabp8622 (2023). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

