A comparative evaluation of computational models for RNA modification detection using nanopore sequencing with RNA004 chemistry
- PMID: 40802798
- PMCID: PMC12346441
- DOI: 10.1093/bib/bbaf404
A comparative evaluation of computational models for RNA modification detection using nanopore sequencing with RNA004 chemistry
Erratum in
-
Correction to: A comparative evaluation of computational models for RNA modification detection using nanopore sequencing with RNA004 chemistry.Brief Bioinform. 2025 Aug 31;26(5):bbaf588. doi: 10.1093/bib/bbaf588. Brief Bioinform. 2025. PMID: 41140068 Free PMC article. No abstract available.
Abstract
Direct RNA sequencing from Oxford Nanopore Technologies has become a valuable method for studying RNA modifications such as N6-methyladenosine (m6A) and pseudouridine (pseU). Recent advancements in the RNA004 chemistry substantially reduce sequencing errors compared to previous chemistries, promising enhanced accuracy for epitranscriptomic analysis. Here we benchmark the performance of two RNA modification detection models for RNA004 data, Dorado and m6Anet, using two wild-type (WT) cell lines (HEK293T and HeLa), with respective ground truths from GLORI and eTAM-seq, and in vitro transcribed (IVT) RNA as negative controls. We found that for m6A sites with ≥10% modification ratio and ≥ 10X coverage, Dorado has higher recall (~0.92) than m6Anet (~0.51). Among true positive predictions, there are high correlations of m6A modification stoichiometry (correlation coefficient of ~0.89 for Dorado-truth and ~ 0.72 for m6Anet-truth). However, combined assessment of WT and IVT datasets show that while the per-site false positive rate can be lower (~8% for Dorado and ~ 33% for m6Anet), both tools can have high per-site false discovery rate of m6A (~40% for Dorado and ~ 80% for m6Anet), or for pseU (~95% for Dorado). Motif analysis reveals that both tools exhibit high heterogeneity of false positive calls across sequence contexts. There is also a substantial overlap of false positive calls between the two IVT samples, suggesting a filtering strategy by compiling a set of low-confidence sites from diverse IVT samples. Our analysis highlights key strengths and limitations of the current generation of m6A detection algorithms and offers insights into optimizing thresholds and interpretability.
Keywords: in vitro transcription; N6-methyladenosine; Oxford Nanopore sequencing; RNA modifications; benchmarking; pseudouridine.
© The Author(s) 2025. Published by Oxford University Press.
Figures
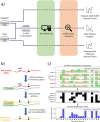

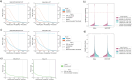
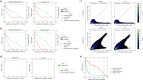

Update of
-
A Comparative Evaluation of Computational Models for RNA modification detection using Nanopore sequencing with RNA004 Chemistry.bioRxiv [Preprint]. 2025 Feb 8:2025.02.03.636352. doi: 10.1101/2025.02.03.636352. bioRxiv. 2025. Update in: Brief Bioinform. 2025 Jul 02;26(4):bbaf404. doi: 10.1093/bib/bbaf404. PMID: 39975272 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

