The immunologic landscape of HRAS-mutant head and neck squamous-cell carcinoma
- PMID: 40803018
- PMCID: PMC12361755
- DOI: 10.1016/j.esmoop.2025.105538
The immunologic landscape of HRAS-mutant head and neck squamous-cell carcinoma
Abstract
Background: HRAS mutations define a distinct biologic subset of head and neck squamous-cell carcinoma (HNSCC). There are limited data regarding HRAS-mutant (mut) tumors' sensitivity to immunotherapy. We sought to evaluate the mutational landscape and transcriptional profile, as well as analyze the tumor microenvironment (TME) of HRAS-mut tumors to provide the conceptual framework for combinatorial treatment approaches.
Materials and methods: We analyzed mutational and transcriptome data from The Cancer Genome Atlas (TCGA). In addition, genomic DNA from baseline tumor biopsies was targeted for sequencing. Our study included 10 patients with HRAS-mut and 40 with HRAS-wild-type (WT) HNSCC. Programmed death-ligand 1 (PD-L1) expression in formalin-fixed paraffin-embedded tumor samples was assessed using the PD-L1 IHC 22C3 pharmDx assay. We characterized subpopulations of exhausted CD8(+) T cells by measuring the expression of T-cell factor-1 (TCF1) and programmed cell death protein 1 (PD-1) in both the center and the periphery of the tumors using multiplex immunohistochemistry, followed by analysis using a manually trained algorithm in QuPath software.
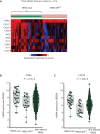
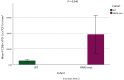
Results: The analysis of TCGA HNSCC mutation and mRNA expression data demonstrated that 6% of HNSCCs harbor mutant HRAS. Transcriptome analysis showed that HRAS-mut HNSCCs are infiltrated by immune cells (CD8A, CD8B, CD2) and have higher expression levels of CXCL11, CXCL10, CXCL9 and CCL4 chemokines. Moreover, the percentage of HRAS-mut samples increased in higher PD-L1 score groups (11% versus 20% versus 100% in tumor positive scores <1%, 1%-49% and ≥50%, respectively, P = 0.006). The analysis of TME showed that HRAS-mut tumors have a statistically significant higher number of total immune cells (5123.17/mm2 versus 3527.93/mm2, P = 0.002) and a higher percentage of pre-exhausted CD8(+) PD-1(+) TCF1(+) T cells in the periphery (384.67/mm2 versus 51.18/mm2, P = 0.040) than HRAS-WT tumors.
Conclusions: HRAS-mut HNSCCs are characterized by a significantly increased number of pre-exhausted PD-1(+) TCF1(+) T cells and PD-L1 expression, suggesting a potential sensitivity to immunotherapy.
Keywords: HRAS mutations; PD-L1 expression; TCF1; head and neck squamous-cell carcinoma; pre-exhausted CD8 (+) T cells.
Copyright © 2025 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures



Similar articles
-
Spatial Transcriptome Analysis of B7-H4 in Head and Neck Squamous Cell Carcinoma: A Novel Therapeutic Target for Anti-Immune Checkpoint Inhibitors.Head Neck Pathol. 2025 Jun 30;19(1):78. doi: 10.1007/s12105-025-01815-w. Head Neck Pathol. 2025. PMID: 40586978 Free PMC article.
-
MHC class I upregulation contributes to the therapeutic response to radiotherapy in combination with anti-PD-L1/anti-TGF-β in squamous cell carcinomas with enhanced CD8 T cell memory-driven response.Cancer Lett. 2025 Jan 1;608:217347. doi: 10.1016/j.canlet.2024.217347. Epub 2024 Nov 22. Cancer Lett. 2025. PMID: 39580046
-
Mapping immune activity in HPV-negative head and neck squamous cell carcinoma: a spatial multiomics analysis.J Immunother Cancer. 2025 Jun 25;13(6):e011851. doi: 10.1136/jitc-2025-011851. J Immunother Cancer. 2025. PMID: 40562703 Free PMC article.
-
Advances and challenges in immunotherapy in head and neck cancer.Front Immunol. 2025 Jun 6;16:1596583. doi: 10.3389/fimmu.2025.1596583. eCollection 2025. Front Immunol. 2025. PMID: 40547025 Free PMC article. Review.
-
A systematic review and meta-analysis of prognostic indicators in patients with head and neck malignancy treated with immune checkpoint inhibitors.J Cancer Res Clin Oncol. 2023 Dec;149(20):18215-18240. doi: 10.1007/s00432-023-05504-5. Epub 2023 Dec 11. J Cancer Res Clin Oncol. 2023. PMID: 38078963 Free PMC article.
References
-
- Mody M.D., Rocco J.W., Yom S.S., Haddad R.I., Saba N.F. Head and neck cancer. Lancet. 2021;398(10318):2289–2299. - PubMed
-
- Mountzios G., Rampias T., Psyrri A. The mutational spectrum of squamous-cell carcinoma of the head and neck: targetable genetic events and clinical impact. Ann Oncol. 2014;25(10):1889–1900. - PubMed
-
- Pearson A.T., Vokes E.E. Is this the dawn of precision oncology in head and neck cancer? J Clin Oncol. 2021;39(17):1839–1841. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

