FedECA: federated external control arms for causal inference with time-to-event data in distributed settings
- PMID: 40804048
- PMCID: PMC12350967
- DOI: 10.1038/s41467-025-62525-z
FedECA: federated external control arms for causal inference with time-to-event data in distributed settings
Abstract
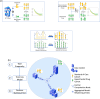
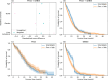
External control arms can inform early clinical development of experimental drugs and provide efficacy evidence for regulatory approval. However, accessing sufficient real-world or historical clinical trials data is challenging. Indeed, regulations protecting patients' rights by strictly controlling data processing make pooling data from multiple sources in a central server often difficult. To address these limitations, we develop a method that leverages federated learning to enable inverse probability of treatment weighting for time-to-event outcomes on separate cohorts without needing to pool data. To showcase its potential, we apply it in different settings of increasing complexity, culminating with a real-world use-case in which our method is used to compare the treatment effect of two approved chemotherapy regimens using data from three separate cohorts of patients with metastatic pancreatic cancer. By sharing our code, we hope it will foster the creation of federated research networks and thus accelerate drug development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the existence of a financial competing interest. Some authors are or were employed by Owkin, Inc. during their time on the project (J. O.d.T, Q.K., M.A., H.L, I.M., N.L., M.H., M.D., T.C., T.F., F.B., J.A.C., Z.Y.). P. L.-P. has received honoraria for consulting and/or advisory board for AMGEN, Pierre Fabre, Biocartis, Servier and BMS. J.B. Bachet has received personal fees from Amgen, Bayer, Bristol Myers Squibb, GlaxoSmithKline, Merck Serono, Merck Sharp & Dohme, Pierre Fabre, Sanofi, Servier, and non-financial support from Amgen, Merck Serono, and Roche, outside the submitted work. J. T. has received honoraria as a speaker and/or in an advisory role from AMGEN, Astelllas, Astra Zeneca, Boehringer, BMS, Merck KGaA, MSD, Novartis, ONO pharmaceuticals, Pierre Fabre, Roche Genentech, Sanofi, Servier and Takeda. A. G. V. has received honoraria as a speaker and/or in an advisory role from Astra Zeneca, Merck Serono, MSD, Novartis, Roche Genentech, Sanofi, and Servier. R. N. has received honoraria as a consultant from Cure51. Part of this work corresponding to work-package 4 of the RHU AI-TRIOMPH and carried out by Owkin France was supported by Agence Nationale de la Recherche as part of the France 2030 plan with reference ANR-23-RHUS-0012 (H.L and F.B.). The remaining authors declare no competing interests.
Figures


Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Taxane monotherapy regimens for the treatment of recurrent epithelial ovarian cancer.Cochrane Database Syst Rev. 2022 Jul 12;7(7):CD008766. doi: 10.1002/14651858.CD008766.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866378 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of topotecan for ovarian cancer.Health Technol Assess. 2001;5(28):1-110. doi: 10.3310/hta5280. Health Technol Assess. 2001. PMID: 11701100
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- DiMasi, J. A., Grabowski, H. G. & Hansen, R. W. Innovation in the pharmaceutical industry: new estimates of r&d costs. J. Health Econ.47, 20–33 (2016). - PubMed
-
- Hay, M., Thomas, D. W., Craighead, J. L., Economides, C. & Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol.32, 40–51 (2014). - PubMed
-
- Dowden, H. & Munro, J. Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov.18, 495–496 (2019). - PubMed
-
- Yin, X. et al. Historic clinical trial external control arm provides an actionable gen-1 efficacy estimate before a randomized trial. JCO Clin. Cancer Inform.7, e2200103 (2023). - PubMed
LinkOut - more resources
Full Text Sources

