The immune-related prognostic gene AIM2 promotes pancreatic cancer progression via inflammasome
- PMID: 40804050
- PMCID: PMC12350923
- DOI: 10.1038/s41598-025-12651-x
The immune-related prognostic gene AIM2 promotes pancreatic cancer progression via inflammasome
Abstract
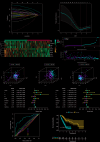
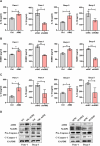
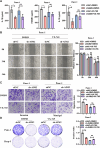
Immune-related factors are closely associated with tumor progression and responses to immunotherapy. A systematic analysis of the immunogenomic landscape and the identification of key immune-related genes (IRGs) can contribute to a better understanding of pancreatic cancer. To identify immune-related genetic prognostic characteristics (IRGPs) of pancreatic cancer, we first constructed an IRGP model containing multiple immune-related genes and determined the relative contributions of each gene using coefficients from the Least Absolute Shrinkage and Selection Operator (LASSO) Cox regression analysis. Subsequently, the prognostic value of the signature was validated through receiver operating characteristic (ROC) curve analysis and Kaplan-Meier survival analysis. Additionally, we explored the potential relationship between IRGPs and immune cell infiltration. Nine gene prognostic features were identified as the optimal IRGPs, which include six high-risk genes and three low-risk genes. Principal Component Analysis (PCA) demonstrated that this feature can effectively distinguish between high-risk and low-risk groups. The area under the curve (AUC) value indicated that IRGPs provide better prognostic clinical utility compared to existing TNM staging classifications. The median overall survival (OS) of high-risk patients was significantly shorter, and the infiltration levels of 24 immune cell types were lower. This study identified nine genes that have been identified as important prognostic biomarkers with immune-related characteristics for pancreatic cancer. Furthermore, we also explored the role of AIM2 in pancreatic cancer among the immune-related signature genes. AIM2 may influence the immune invasion and immunotherapy of pancreatic cancer by promoting the inflammatory environment of pancreatic cancer. AIM2 could be a new therapeutic target for pancreatic cancer.
Keywords: AIM2; Immune infiltration; Immune-related genes; Inflammasome; Pancreatic cancer; Prognostic signature; Tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Systematic Analysis of an Immune-Related Gene Signature for Predicting Prognosis and Immune Characteristics in Primary Lower Grade Glioma.Biomed Res Int. 2025 Aug 12;2025:6180391. doi: 10.1155/bmri/6180391. eCollection 2025. Biomed Res Int. 2025. PMID: 40838069 Free PMC article.
-
A novel copper-induced cell death-related lncRNA prognostic signature associated with immune infiltration and clinical value in gastric cancer.J Cancer Res Clin Oncol. 2023 Sep;149(12):10543-10559. doi: 10.1007/s00432-023-04916-7. Epub 2023 Jun 8. J Cancer Res Clin Oncol. 2023. PMID: 37291405 Free PMC article.
-
TGFB2 Gene Methylation in Tumors with Low CD8+ T-Cell Infiltration Drives Positive Prognostic Overall Survival Responses in Pancreatic Ductal Adenocarcinoma.Int J Mol Sci. 2025 Jun 10;26(12):5567. doi: 10.3390/ijms26125567. Int J Mol Sci. 2025. PMID: 40565031 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
-
- Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin.71, 7–33 (2021). - PubMed
-
- Karamitopoulou, E. Emerging prognostic and predictive factors in pancreatic Cancer. Mod. Pathol.36, 100328 (2023). - PubMed
-
- Balaban, E. P., Mangu, P. B. & Yee, N. S. Locally advanced unresectable pancreatic cancer: American society of clinical oncology clinical practice guideline summary. J. Oncol. Pract.13, 265–269 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

