Multidimensional pan cancer analysis of the sodium induced cell death gene TRPM4
- PMID: 40804073
- PMCID: PMC12350802
- DOI: 10.1038/s41598-025-15082-w
Multidimensional pan cancer analysis of the sodium induced cell death gene TRPM4
Abstract
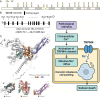
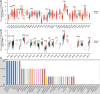
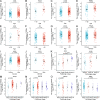
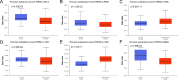
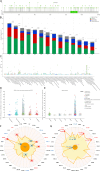
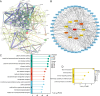
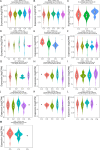
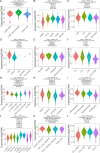
Cell death modalities play crucial roles in cancer evolution and therapeutic responses. Among various mechanisms, necrosis by sodium overload (NECSO) is a newly recognized process initiated by disruptions in Na+ homeostasis, manifesting through osmotic stress, energy depletion, and immunogenic damage. The TRPM4 gene, which encodes a calcium-activated and sodium-selective ion channel, has surfaced as a significant regulator connecting ionic metabolism with oncogenic pathways. Given these insights, our study aims to comprehensively analyze the expression and implications of TRPM4 across diverse cancer types to elucidate its potential as a biomarker and therapeutic target. We conducted a systematic investigation of TRPM4 across 33 cancer types defined by the Cancer Genome Atlas (TCGA), integrating transcriptomic, proteomic, epigenetic, and clinical datasets from TCGA, GTEx, and Human Protein Atlas (HPA). We employed differential expression analyses, receiver operating characteristic (ROC) curves, and survival analyses, alongside mutation and methylation assessments. Furthermore, we explored TRPM4's immunological aspects through immune infiltration analyses. Our analyses revealed significant TRPM4 overexpression in several tumors, such as bladder (BLCA), cholangiocarcinoma (CHOL), and ovarian cancer (OV), whilst being downregulated in others like kidney clear cell carcinoma (KIRC) and lung adenocarcinoma (LUAD). Notably, TRPM4 expression correlated with overall survival, disease-specific survival, and progression-free interval, highlighting its prognostic value. Furthermore, promoter methylation and mutation patterns elucidated the mechanisms underlying TRPM4 dysregulation, and immune infiltration analyses suggested its involvement in tumor immune evasion. This investigation highlights TRPM4's dual role in mediating sodium-induced cell death and modulating the tumor microenvironment, proposing it as a potential biomarker for cancer diagnosis and prognosis, though its association with demographic and pathological characteristics appears limited and tumor-type specific. Given its import in various malignancies and potential therapeutic implications through ion channel-focused strategies, TRPM4 warrants further exploration as a target for precision oncology.
Keywords: Immune evasion; Ion channels; Pan-cancer analysis; Sodium-induced cell death (NECSO); TRPM4; Tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures







References
-
- Balaji, S., Terrero, D., Tiwari, A. K., Ashby, C. R. Jr. & Raman, D. Alternative approaches to overcome chemoresistance to apoptosis in cancer. Adv. Protein Chem. Struct. Biol.126, 91–122. 10.1016/bs.apcsb.2021.01.005 (2021). - PubMed
-
- AbdulHussein, A. H. et al. Mechanisms of cancer cell death induction by triptolide. BioFactors49, 718–735. 10.1002/biof.1944 (2023). - PubMed
-
- Cai, J., Xu, X. & Saw, P. E. Nanomedicine targeting ferroptosis to overcome anticancer therapeutic resistance. Sci. China Life Sci.67, 19–40. 10.1007/s11427-022-2340-4 (2024). - PubMed
-
- Fu, W. et al. Persistent activation of TRPM4 triggers necrotic cell death characterized by sodium overload. Nat. Chem. Biol.10.1038/s41589-025-01841-3 (2025). - PubMed
-
- Rajamanickam, G., Hu, Z. & Liao, P. Targeting the TRPM4 channel for neurologic diseases: Opportunity and challenge. Neuroscientist10.1177/10738584251318979 (2025). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

