Methylation-induced suppression of BEX1 activates AKT/ERK/STAT3 signaling pathways regulating cell cycle and apoptosis in glioma
- PMID: 40804095
- PMCID: PMC12350733
- DOI: 10.1038/s41598-025-14123-8
Methylation-induced suppression of BEX1 activates AKT/ERK/STAT3 signaling pathways regulating cell cycle and apoptosis in glioma
Abstract
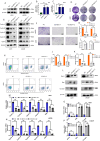
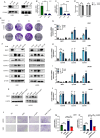
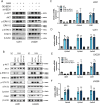
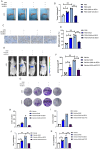
Gliomas are highly aggressive brain tumors with complex molecular characteristics. The role of BEX1 methylation in gliomas is not well understood, despite its potential implications for tumor biology and therapy. Investigating this relationship could uncover critical mechanisms underlying glioma pathogenesis and highlight therapeutic targets. This study aims to elucidate the specific mechanisms by which BEX1 methylation regulates the cell cycle and apoptosis in glioma. We conducted bioinformatics analyses to assess BEX1 expression differences in glioma using tissue samples, followed by validation through Western blot and qRT-PCR. Functional assays in glioma cell lines were performed, employing gene transfection and small molecule inhibitors to further explore BEX1's role in the AKT/ERK/STAT3 signaling pathways. Our findings reveal that BEX1 is significantly downregulated in gliomas due to promoter methylation, which in turn activates the AKT/ERK/STAT3 signaling cascade, leading to alterations in cell cycle regulation and apoptosis. Targeting BEX1 methylation presents a promising therapeutic avenue for glioma treatment. This study provides valuable insights into the mechanisms of BEX1 in glioma, paving the way for clinical translation and further research.
Keywords: BEX1; Clinical translation; Giloma; Methylation; PI3K/ERK/STAT3.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Integrin αVβ1-activated PYK2 promotes the progression of non-small-cell lung cancer via the STAT3-VGF axis.Cell Commun Signal. 2024 Jun 6;22(1):313. doi: 10.1186/s12964-024-01639-1. Cell Commun Signal. 2024. PMID: 38844957 Free PMC article.
-
Systematic Review of Molecular Targeted Therapies for Adult-Type Diffuse Glioma: An Analysis of Clinical and Laboratory Studies.Int J Mol Sci. 2023 Jun 21;24(13):10456. doi: 10.3390/ijms241310456. Int J Mol Sci. 2023. PMID: 37445633 Free PMC article.
-
Construction and validation of a prognostic model for glioma: an analysis based on mismatch repair-related genes and their correlation with clinicopathological features.Transl Cancer Res. 2025 May 30;14(5):2690-2706. doi: 10.21037/tcr-24-2045. Epub 2025 May 9. Transl Cancer Res. 2025. PMID: 40530129 Free PMC article.
-
Remote Ischemic Postconditioning Improve Cerebral Ischemia-Reperfusion Injury Induced Cognitive Dysfunction through Suppressing Mitochondrial Apoptosis in Hippocampus via TK/BK/B2R-Mediated PI3K/AKT.Mol Neurobiol. 2025 Aug;62(8):10652-10669. doi: 10.1007/s12035-025-04864-y. Epub 2025 Apr 14. Mol Neurobiol. 2025. PMID: 40229456 Free PMC article.
-
Glioma Stem Cells as Promoter of Glioma Progression: A Systematic Review of Molecular Pathways and Targeted Therapies.Int J Mol Sci. 2024 Jul 22;25(14):7979. doi: 10.3390/ijms25147979. Int J Mol Sci. 2024. PMID: 39063221 Free PMC article.
References
-
- Lim, M., Xia, Y., Bettegowda, C. & Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol.15, 422–442. 10.1038/s41571-018-0003-5 (2018). - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

