In vivo crosslinking and effective 2D enrichment for proteome wide interactome studies
- PMID: 40804100
- PMCID: PMC12350791
- DOI: 10.1038/s42004-025-01644-6
In vivo crosslinking and effective 2D enrichment for proteome wide interactome studies
Abstract
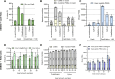
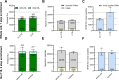
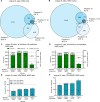
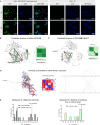
Cross-linking mass spectrometry has evolved as a powerful technique to study protein-protein interactions and to provide structural information. Low reaction efficiencies, and complex matrices lead to challenging system wide crosslink analysis. We improved and streamlined an Azide-A-DSBSO based in vivo crosslinking workflow employing two orthogonal effective enrichment steps: Affinity enrichment and size exclusion chromatography (SEC). Combined, they allow an effective enrichment of DSBSO containing peptides and remove the background of linear as well as mono-linked peptides. We found that the analysis of a single SEC fraction is effective to yield ~90% of all crosslinks, which is important whenever measurement time is limited, and sample throughput is crucial. Our workflow resulted in more than 5000 crosslinks from K562 cells and generated a comprehensive PPI network. From 393 PPI found within the nucleus, 56 are novel. We further show, that by applying DSBSO to nuclear extracts we yield more crosslinks on lower abundant proteins and showcase this on the DEAD-box RNA helicase DDX39B which is predominantly expressed in the nucleus. Our data indicates that DDX39B might be present in monomeric and dimeric forms together with DDX39A within the nuclear extracts analyzed.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Iacobucci, C., Götze, M. & Sinz, A. Cross-linking/mass spectrometry to get a closer view on protein interaction networks. Curr. Opin. Biotechnol.63, 48–53 (2020). - PubMed
-
- Sinz, A. Divide and conquer: cleavable cross-linkers to study protein conformation and protein–protein interactions. Anal. Bioanal. Chem.409, 33–44 (2017). - PubMed
-
- Steigenberger, B., Albanese, P., Heck, A. J. R. & Scheltema, R. A. To cleave or not to cleave in XL-MS? J. Am. Soc. Mass Spectrom. 10.1021/jasms.9b00085 (2019). - PubMed
Grants and funding
- LS20-079/Vienna Science and Technology Fund (Wiener Wissenschafts-, Forschungs- und Technologiefonds)
- 10.55776/P35045/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- 10.55776/F88/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- DOI 10.55776/ESP566/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- 4795911/Österreichische Forschungsförderungsgesellschaft (Austrian Research Promotion Agency)
LinkOut - more resources
Full Text Sources

