PharmaFormer predicts clinical drug responses through transfer learning guided by patient derived organoid
- PMID: 40804292
- PMCID: PMC12350944
- DOI: 10.1038/s41698-025-01082-6
PharmaFormer predicts clinical drug responses through transfer learning guided by patient derived organoid
Abstract
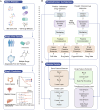
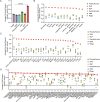
A major challenge in effective cancer treatment is the variability of drug responses among patients. Patient-derived organoids greatly preserve the genetic and histological characteristics even the drug sensitivities of primary tumor tissues, therefore provide a compelling approach to predict clinical outcome. However, the individual organoid culture and following drug response test are time and cost-consuming, which hinders the potential clinical application. Here, we developed PharmaFormer, a clinical drug response prediction model based on custom Transformer architecture and transfer learning. PharmaFormer was initially pre-trained with the abundant gene expression and drug sensitivity data of 2D cell lines, and was then finalized through a model further fine-tuned with the limited organoid pharmacogenomic data accumulated at the present stage. Our results demonstrate that PharmaFormer, integrating both pan-cancer cell lines and organoids of a specific type of tumor, provides a dramatically improved accurate prediction of clinical drug response. This study highlights that advanced AI models combined with biomimetic organoid models will accelerate precision medicine and future drug development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Organoid Models Established from Primary Tumors and Patient-Derived Xenograft Tumors Reflect Platinum Sensitivity of Ovarian Cancer Patients.bioRxiv [Preprint]. 2025 May 2:2024.06.28.601283. doi: 10.1101/2024.06.28.601283. bioRxiv. 2025. PMID: 40654830 Free PMC article. Preprint.
-
Interplay between tumor mutation burden and the tumor microenvironment predicts the prognosis of pan-cancer anti-PD-1/PD-L1 therapy.Front Immunol. 2025 Jul 24;16:1557461. doi: 10.3389/fimmu.2025.1557461. eCollection 2025. Front Immunol. 2025. PMID: 40777041 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of topotecan for ovarian cancer.Health Technol Assess. 2001;5(28):1-110. doi: 10.3310/hta5280. Health Technol Assess. 2001. PMID: 11701100
References
-
- Foulkes, W. D., Smith, I. E. & Reis-Filho, J. S. Triple-negative breast cancer. N. Engl. J. Med.363, 1938–1948 (2010). - PubMed
-
- Bruix, J. et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet389, 56–66 (2017). - PubMed
-
- Lopez-Beltran, A. et al. Advances in diagnosis and treatment of bladder cancer. BMJ384, e076743 (2024). - PubMed
-
- Guo, L. et al. Molecular profiling provides clinical insights into targeted and immunotherapies as well as colorectal cancer prognosis. Gastroenterology165, 414–428.e7 (2023). - PubMed
LinkOut - more resources
Full Text Sources

