GenMasterTable: a user-friendly desktop application for filtering, summarising, and visualising large-scale annotated genetic variants
- PMID: 40804356
- PMCID: PMC12344886
- DOI: 10.1186/s12859-025-06238-6
GenMasterTable: a user-friendly desktop application for filtering, summarising, and visualising large-scale annotated genetic variants
Abstract
Background: The rapid expansion of next-generation sequencing (NGS) technologies has generated vast amounts of genomic data, creating a growing demand for secure, scalable, and accessible tools to support variant interpretation. However, many existing solutions are command-line based, rely on cloud or server infrastructures that may pose data privacy risks, lack flexibility in supporting both VCF, CSV and TSV formats, or struggle to handle the scale and complexity of modern genomic datasets. There is a clear need for a user-friendly, locally operated application capable of efficiently processing annotated variant data for large-scale cohort level analysis.
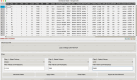
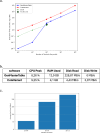
Results: We introduce GenMasterTable, a free, secure, and cross-platform desktop application designed to simplify variant analysis through an intuitive graphical user interface (GUI). As the first tool to enable comprehensive cohort-level analysis from VCF, CSV to TSV files, GenMasterTable provides advanced functionality for concatenation, filtering, summarizing, and visualizing large-scale annotated datasets. Tailored for users without programming expertise, it enables rapid and accurate exploration of genetic variants, making it a practical solution for both research and clinical settings.
Conclusion: GenMasterTable addresses critical limitations in current variant analysis workflows by combining usability, data security, and scalability. Its support for multiple input formats and locally executed operations empowers clinicians, geneticists, and researchers to perform comprehensive variant analysis efficiently without the need for programming expertise.
Supplementary Information: The online version contains supplementary material available at 10.1186/s12859-025-06238-6.
Keywords: Cohort-level analysis; Desktop bioinformatics tool; Genetic variant analysis; Graphical user interface (GUI); Next-generation sequencing (NGS).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials

