Structural and functional roles of conserved residues of human papillomavirus (HPV) E2 protein and biological consequences
- PMID: 40804408
- PMCID: PMC12345011
- DOI: 10.1186/s12985-025-02903-7
Structural and functional roles of conserved residues of human papillomavirus (HPV) E2 protein and biological consequences
Abstract
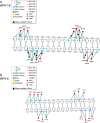
Background: Human papillomavirus (HPV) is a prevalent viral pathogen that causes a variety of malignancies, including cervical cancer, one of the leading causes of cancer-related deaths among women worldwide. The HPV E2 protein is a central regulator of viral replication and oncogene expression, making it a critical determinant of HPV-associated malignancies. While its core functions are conserved, variations within the E2 protein are thought to contribute to the differential oncogenic potential among HPV types, though the structural basis for this remains incompletely understood. Previous research from our laboratory suggests that mutations within a 12-base pair segment of the long control region that encompasses the E2 binding sites may influence the oncogenic potential of certain HPV strains.
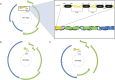
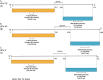
Methods: Computational methods, including multiple sequence alignment, phylogenetic analysis, and protein structural modeling were employed to identify conserved regions and correlate these with potential cancer-associated mutations in the coding region.
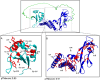
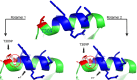
Results: Structural modeling using AlphaFold3 and visualization in PyMOL revealed that conserved E2 residues cluster near the DNA-binding surface in the C-terminal domain and at critical interaction sites in the N-terminal transactivation domain, important for E1 DNA helicase binding and potentially other host factor interactions. Notably, species-specific adaptations, including the T309P substitution in the HPV52 subfamily B2, which may induce structural changes in the DNA-binding domain, and variations in the 12-base pair spacer, could modulate oncogene expression.
Conclusions: Collectively, these findings refine our understanding of E2's essential role in viral pathogenesis and highlight promising targets for therapeutic intervention in high-risk HPV strains.
Keywords: DNA-binding domain; E2 protein; Human papillomavirus; Oncogenic potential; Sequence conservation; Structural modeling; T309P mutation; Transactivation domain.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: All authors have explicitly authorized this publication. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Factors that influence caregivers' and adolescents' views and practices regarding human papillomavirus (HPV) vaccination for adolescents: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2025 Apr 15;4(4):CD013430. doi: 10.1002/14651858.CD013430.pub2. Cochrane Database Syst Rev. 2025. PMID: 40232221 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
A conserved cysteine in the DNA-binding domain of MmuPV1 E2 is required for replication in vivo.J Virol. 2025 Jan 31;99(1):e0142324. doi: 10.1128/jvi.01423-24. Epub 2024 Dec 12. J Virol. 2025. PMID: 39665560 Free PMC article.
-
Targeting papillomavirus infections: high-throughput screening reveals an effective inhibitor of cutaneous β-HPV types.J Virol. 2025 Aug 19;99(8):e0091825. doi: 10.1128/jvi.00918-25. Epub 2025 Jul 8. J Virol. 2025. PMID: 40626718 Free PMC article.
-
Interventions targeted at women to encourage the uptake of cervical screening.Cochrane Database Syst Rev. 2021 Sep 6;9(9):CD002834. doi: 10.1002/14651858.CD002834.pub3. Cochrane Database Syst Rev. 2021. PMID: 34694000 Free PMC article.
References
-
- Han F, Guo X, ying, Jiang M, xia, Xia Nshao, Gu Y, Li S. wei. Structural biology of the human papillomavirus. Structure. 2024;32(11):1877–92. - PubMed
LinkOut - more resources
Full Text Sources

