DNA methylation variations of DNA damage response in glioblastoma: NSUN5 modulates tumor-intrinsic cytosolic DNA-sensing and microglial behavior
- PMID: 40804410
- PMCID: PMC12351926
- DOI: 10.1186/s12967-025-06360-2
DNA methylation variations of DNA damage response in glioblastoma: NSUN5 modulates tumor-intrinsic cytosolic DNA-sensing and microglial behavior
Abstract
Background: Variations in DNA methylation within the DNA damage response (DDR) mechanism could have significant implications for glioma prognosis and immune responses. This study aimed to explore the global DNA methylation landscape of DDR genes in gliomas and identify key epigenetically regulated genes influencing glioma biology and immunity.
Methods: This study incorporated a range of public and local glioma datasets. Multiple clinical, bioinformatic, and in vitro experimental analyses were conducted to explore clinical and biological aspects.
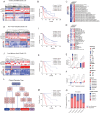
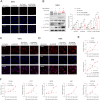
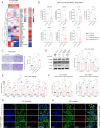
Results: Global DNA methylation variations in DDR genes correlated with distinct glioma prognoses, with five CpGs identified as potent predictors. Hierarchical clustering and a risk-score model based on these CpGs unveiled immune-related prognostic subgroups in glioblastomas (GBMs) and lower-grade gliomas (LGGs). NSUN5, epigenetically regulated by one of these CpGs, highlighted the biological significance of the DDR CpG panel. In vitro, NSUN5 displayed tumor-suppressor-like activities in GBM cells, but clinically, it was an unfavorable prognostic marker. Depletion of NSUN5 shifted cytosolic DNA sensing from a STING-dependent (cGAS-STING) pathway to a STING-independent (DNA-PK-HSPA8) pathway, leading to a delayed but more robust type I interferon (IFN) response in GBM cells and enhancing microglial M1 polarization and chemotaxis. This may partially account for the functional discrepancy of NSUN5 observed between experimental and clinical contexts.
Conclusion: This study highlights the complex interplay between DNA methylation, the DDR mechanism, cytosolic DNA sensing, and glioma immunity. These findings may inspire novel strategies for DNA sensing-based immunotherapy.
Keywords: NSUN5; Cytosolic DNA sensing; DNA damage response; DNA methylation; Glioma.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Informed consent was obtained from all participants from the Neurosurgery Departments of Rennes and Angers University Hospitals and the Department of Neurosurgery, Xijing Hospital. This study was approved by the Institutional Review Board at Xijing Hospital, Air Force Medical University (No. KY20223039-1). Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures




Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Magnetic resonance perfusion for differentiating low-grade from high-grade gliomas at first presentation.Cochrane Database Syst Rev. 2018 Jan 22;1(1):CD011551. doi: 10.1002/14651858.CD011551.pub2. Cochrane Database Syst Rev. 2018. PMID: 29357120 Free PMC article.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Antiangiogenic therapy for high-grade glioma.Cochrane Database Syst Rev. 2014 Sep 22;(9):CD008218. doi: 10.1002/14651858.CD008218.pub3. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2018 Nov 22;11:CD008218. doi: 10.1002/14651858.CD008218.pub4. PMID: 25242542 Updated.
References
-
- Kornepati A, Rogers CM, Sung P, Curiel TJ. The complementarity of Ddr, nucleic acids and anti-tumour immunity. Nature. 2023;619(7970):475–86. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

