Multi-omics analysis of bidirectional liquid-liquid phase separation signatures reveals immunotherapy response and prognosis in renal cell carcinoma
- PMID: 40804718
- PMCID: PMC12351944
- DOI: 10.1186/s12885-025-14749-x
Multi-omics analysis of bidirectional liquid-liquid phase separation signatures reveals immunotherapy response and prognosis in renal cell carcinoma
Abstract
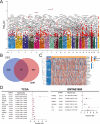
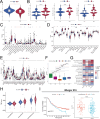
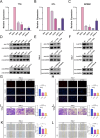
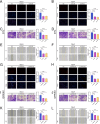
Renal cell carcinoma (RCC) is a common malignant tumor of the urinary system, accounting for approximately 80% of urinary tract malignancies, with clear cell renal cell carcinoma (ccRCC) being the most prevalent subtype. Despite its high incidence, the pathogenesis of ccRCC remains incompletely elucidated, and limitations persist in early diagnosis, targeted therapy, and immunotherapy. Based on emerging evidence highlighting the critical role of liquid-liquid phase separation (LLPS) in tumor proliferation and invasion, this study leveraged multiple ccRCC cohorts to employ Mendelian randomization (MR) and ultimately developed a novel Bidirectional Liquid-Liquid Phase Separation-associated Index (B-LLPSI) using LASSO regression, aiming to predict patient prognosis and therapeutic response. Notably, B-LLPSI demonstrates substantial potential as a universal prognostic indicator across multiple cancer types, underpinning its prospective application in individualized oncology. Paradoxically, while high B-LLPSI levels correlate with poorer patient prognosis, they simultaneously predict enhanced responsiveness to immunotherapy. Furthermore, the B-LLPSI signature was found to be upregulated in tumor tissues and capable of promoting tumor proliferation and invasion. In summary, B-LLPSI holds promise as a robust and independent prognostic tool for diverse cancers, providing critical insights for formulating individualized treatment strategies.
Keywords: Immune microenvironment; Immunotherapy efficient; Liquid-liquid phase separation; Molecular docking; Pancancer analysis; Prognosis; Renal clear cell carcinoma.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Consent for publication: Not applicable. Conflict of interest: The authors declare no conflicts of interest. Not applicable.
Figures




Similar articles
-
COL6A2 in clear cell renal cell carcinoma: a multifaceted driver of tumor progression, immune evasion, and drug sensitivity.J Transl Med. 2025 Aug 6;23(1):875. doi: 10.1186/s12967-025-06793-9. J Transl Med. 2025. PMID: 40770761 Free PMC article.
-
NME4: A novel metabolic-associated biomarker for prognosis prediction and immunotherapy response evaluation in clear cell renal cell carcinoma.Mol Immunol. 2025 Aug;184:149-163. doi: 10.1016/j.molimm.2025.06.011. Epub 2025 Jul 1. Mol Immunol. 2025. PMID: 40602220
-
Multi-omics analysis reveals the role of ribosome biogenesis in malignant clear cell renal cell carcinoma and the development of a machine learning-based prognostic model.Front Immunol. 2025 Jun 26;16:1602898. doi: 10.3389/fimmu.2025.1602898. eCollection 2025. Front Immunol. 2025. PMID: 40642093 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
A Systematic Review and Meta-analysis Comparing the Effectiveness and Adverse Effects of Different Systemic Treatments for Non-clear Cell Renal Cell Carcinoma.Eur Urol. 2017 Mar;71(3):426-436. doi: 10.1016/j.eururo.2016.11.020. Epub 2016 Dec 8. Eur Urol. 2017. PMID: 27939075
References
-
- Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67:507–24. 10.3322/caac.21411. - PubMed
-
- Chakraborty S, Mishra J, Roy A, Niharika N, Manna S, Baral T, Nandi P, Patra S, Patra SK. Liquid-liquid phase separation in subcellular assemblages and signaling pathways: chromatin modifications induced gene regulation for cellular physiology and functions including carcinogenesis. Biochimie S. 2024;00101–9. 10.1016/j.biochi.2024.05.007. 0300-9084(24). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

