Optimized Ribonucleoprotein Complexes Enhance Prime Editing Efficiency in Zebrafish
- PMID: 40805085
- PMCID: PMC12345536
- DOI: 10.3390/ani15152295
Optimized Ribonucleoprotein Complexes Enhance Prime Editing Efficiency in Zebrafish
Abstract
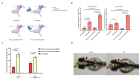
Prime editing (PE) has emerged as a transformative genome editing technology, enabling precise base substitutions, insertions, and deletions without inducing double-strand DNA breaks (DSBs). However, its application in zebrafish remains limited by low efficiency. Here, we leveraged PE7, a state-of-the-art PE system, combined with La-accessible prime editing guide RNAs (pegRNAs), to enhance editing efficiency in zebrafish. By co-incubating PE7 protein with La-accessible pegRNAs to form ribonucleoprotein (RNP) complexes and microinjecting these complexes into zebrafish embryos, we achieved up to 15.99% editing efficiency at target loci-an improvement of 6.81- to 11.46-fold over PE2. Additionally, we observed 16.60% 6 bp insertions and 13.18% 10 bp deletions at the adgrf3b locus, representing a 3.13-fold increase over PE2. Finally, we used PE to introduce desired edits at the tyr locus, successfully generating zebrafish with the tyr P302L mutation that exhibited melanin reduction. These findings demonstrate that PE7 significantly enhances prime editing efficiency in fish, providing novel tools for functional gene studies and genetic breeding in aquatic species.
Keywords: aquaculture; genome editing; prime editor; zebrafish.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

