The Kinase Inhibitor GNF-7 Is Synthetically Lethal in Topoisomerase 1-Deficient Ewing Sarcoma
- PMID: 40805174
- PMCID: PMC12346386
- DOI: 10.3390/cancers17152475
The Kinase Inhibitor GNF-7 Is Synthetically Lethal in Topoisomerase 1-Deficient Ewing Sarcoma
Abstract
Background/objectives: Ewing sarcoma (ES), a highly aggressive bone and soft tissue cancer occurring in children and young adults, is defined by the ETS fusion oncoprotein EWS::FLI1. Although event-free survival rates remain high in ES patients with localized disease, those with metastatic or relapsed disease face poor long-term survival odds. Topoisomerase 1 (TOP1) inhibitors are commonly used therapeutics in ES relapse regimens.
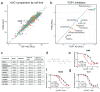
Methods: In this work, we used a genome-wide CRISPR knockout library screen to identify the deletion of the TOP1 gene as a mechanism for resistance to topoisomerase 1 inhibitors. Using isogenic cell line models, we performed a high-throughput small-molecule screen to discover a small molecule, GNF-7, which had an IC50 that was 10-fold lower in TOP1-deficient cells when compared to the wild-type cells.
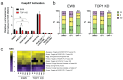
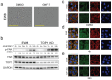
Results: The characterization of GNF-7 demonstrated the molecule was highly active in the inhibition of CSK, p38α, EphA2, Lyn, and ZAK and specifically downregulated genes induced by the EWS::FLI1 fusion oncoprotein.
Conclusions: Together, these results suggest that GNF-7 or small molecules with a similar kinase profile could be effective treatments for ES patients in combination with TOP1 inhibitors or for those patients who have developed resistance to TOP1 inhibitors.
Keywords: Ewing sarcoma; GNF-7; focal adhesion; irinotecan; kinase inhibitor; topoisomerase.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Kolb E.A., Kushner B.H., Gorlick R., Laverdiere C., Healey J.H., LaQuaglia M.P., Huvos A.G., Qin J., Vu H.T., Wexler L., et al. Long-term event-free survival after intensive chemotherapy for Ewing’s family of tumors in children and young adults. J. Clin. Oncol. 2003;21:3423–3430. doi: 10.1200/JCO.2003.10.033. - DOI - PubMed
-
- Gupta A., Dietz M.S., Riedel R.F., Dhir A., Borinstein S.C., Isakoff M.S., Aye J.M., Rainusso N., Armstrong A.E., DuBois S.G., et al. Consensus recommendations for systemic therapies in the management of relapsed Ewing sarcoma: A report from the National Ewing Sarcoma Tumor Board. Cancer. 2024;130:4028–4039. doi: 10.1002/cncr.35537. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

