Ribosome Biogenesis and Function in Cancer: From Mechanisms to Therapy
- PMID: 40805230
- PMCID: PMC12346075
- DOI: 10.3390/cancers17152534
Ribosome Biogenesis and Function in Cancer: From Mechanisms to Therapy
Abstract
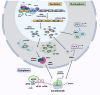
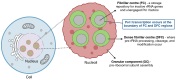
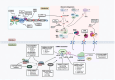
Ribosome biogenesis is a highly coordinated, multi-step process that assembles the ribosomal machinery responsible for translating mRNAs into proteins. It begins with the rate-limiting step of RNA polymerase I (Pol I) transcription of the 47S ribosomal RNA (rRNA) genes within a specialised nucleolar region in the nucleus, followed by rRNA processing, modification, and assembly with ribosomal proteins and the 5S rRNA produced by Pol III. The ribosomal subunits are then exported to the cytoplasm to form functional ribosomes. This process is tightly regulated by the PI3K/RAS/MYC oncogenic network, which is frequently deregulated in many cancers. As a result, ribosome synthesis, mRNA translation, and protein synthesis rates are increased. Growing evidence supports the notion that dysregulation of ribosome biogenesis and mRNA translation plays a pivotal role in the pathogenesis of cancer, positioning the ribosome as a promising therapeutic target. In this review, we summarise current understanding of dysregulated ribosome biogenesis and function in cancer, evaluate the clinical development of ribosome targeting therapies, and explore emerging targets for therapeutic intervention in this rapidly evolving field.
Keywords: cancer therapy; mRNA translation; nucleolus; pol I transcription; ribosome biogenesis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Focal adhesion kinase promotes ribosome biogenesis to drive advanced thyroid cancer cell growth and survival.Front Oncol. 2025 May 19;15:1252544. doi: 10.3389/fonc.2025.1252544. eCollection 2025. Front Oncol. 2025. PMID: 40458728 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Phase separation via protein-protein and protein-RNA networks coordinates ribosome assembly in the nucleolus.Biochim Biophys Acta Gen Subj. 2025 Sep;1869(10):130835. doi: 10.1016/j.bbagen.2025.130835. Epub 2025 Jul 16. Biochim Biophys Acta Gen Subj. 2025. PMID: 40675030 Review.
-
Explainable machine learning reveals ribosome biogenesis biomarkers in preeclampsia risk prediction.Front Immunol. 2025 Jun 9;16:1595222. doi: 10.3389/fimmu.2025.1595222. eCollection 2025. Front Immunol. 2025. PMID: 40552285 Free PMC article.
-
Nucleolar Organization in Response to Transcriptional Stress.Cancer Sci. 2025 Jul 29. doi: 10.1111/cas.70164. Online ahead of print. Cancer Sci. 2025. PMID: 40726293 Review.
References
-
- Unuma T., Busch H. Formation of microspherules in nucleoli of tumor cells treated with high doses of actinomycin D. Cancer Res. 1967;27:1232–1242. - PubMed
-
- Daskal Y., Woodard C., Crooke S.T., Busch H. Comparative ultrastructural studies of nucleoli of tumor cells treated with adriamycin and the newer anthracyclines, carminomycin and marcellomycin. Cancer Res. 1978;38:467–473. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

