Transcriptome Analysis and Functional Characterization of the HvLRR_8-1 Gene Involved in Barley Resistance to Pyrenophora graminea
- PMID: 40805699
- PMCID: PMC12348818
- DOI: 10.3390/plants14152350
Transcriptome Analysis and Functional Characterization of the HvLRR_8-1 Gene Involved in Barley Resistance to Pyrenophora graminea
Abstract
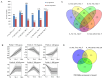
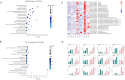
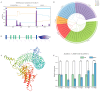
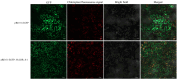
Barley leaf stripe, caused by Pyrenophora graminea (Pg), significantly reduces yields across various regions globally. Understanding the resistance mechanisms of barley to Pg is crucial for advancing disease resistance breeding efforts. In this study, two barley genotypes-highly susceptible Alexis and immune Ganpi2-were inoculated with the highly pathogenic Pg isolate QWC for 7, 14, and 18 days. The number of differentially expressed genes (DEGs) in Alexis was 1350, 1898, and 2055 at 7, 14, and 18 days, respectively, while Ganpi2 exhibited 1195, 1682, and 2225 DEGs at the same time points. Gene expression pattern analysis revealed that Alexis responded more slowly to Pg infection compared to Ganpi2. A comparative analysis identified 457 DEGs associated with Ganpi2's immunity to Pg. Functional enrichment of these DEGs highlighted the involvement of genes related to plant-pathogen interactions and kinase activity in Pg immunity. Additionally, 20 resistance genes and 24 transcription factor genes were predicted from the 457 DEGs. Twelve candidate genes were selected for qRT-PCR verification, and the results showed that the transcriptomic data was reliable. We conducted cloning of the candidate Pg resistance gene HvLRR_8-1 by the barley cultivar Ganpi2, and the sequence analysis confirmed that the HvLRR_8-1 gene contains seven leucine-rich repeat (LRR) domains and an S_TKc domain. Subcellular localization in tobacco indicates that the HvLRR_8-1 is localized on the cell membrane. Through the functional analysis using virus-induced gene silencing, it was demonstrated that HvLRR_8-1 plays a critical role in regulating barley resistance to Pg. This study represents the first comparative transcriptome analysis of barley varieties with differing responses to Pg infection, providing that HvLRR_8-1 represents a promising candidate gene for improving durable resistance against Pg in cultivated barley.
Keywords: BSMV-VIGS; Hordeum vulgare L.; HvLRR_8-1; Pyrenophora graminea; barley leaf stripe; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest regarding this article.
Figures

Similar articles
-
Mutagenesis of Highland barley (Hordeum vulgare L. Var. nudum) using nitrogen ion beam implantation: screening of phenotypic Var.ations and comparative transcriptome analysis.BMC Genomics. 2025 Jul 21;26(1):681. doi: 10.1186/s12864-025-11856-8. BMC Genomics. 2025. PMID: 40691530 Free PMC article.
-
Comparative transcriptome analysis reveals the potential mechanism of seed germination promoted by trametenolic acid in Gastrodia elata Blume.Sci Rep. 2025 Jul 24;15(1):26869. doi: 10.1038/s41598-025-12269-z. Sci Rep. 2025. PMID: 40702084 Free PMC article.
-
Identification of new candidate genes affecting drip loss in pigs based on genomics and transcriptomics data.J Anim Sci. 2025 Jan 4;103:skaf177. doi: 10.1093/jas/skaf177. J Anim Sci. 2025. PMID: 40485044 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
References
-
- Valè G., Tacconi G., Francia E., Dall’Aglio E., Govoni C., Pecchioni N., Arru L., Delogu G., Porta-Puglia A., Stanca A. Advances in understanding barley Pyrenophora graminea interactions; Proceedings of the 2nd International Workshop on Barley Leaf Blights; Aleppo, Syria. 7–11 April 2002.
-
- Bulgarelli D., Boselli C., Collins N.C., Consonni G., Stanca A.M., Schulzelefert P., Valè G. The CC-NB-LRR-type Rdg2a resistance gene confers immunity to the seed-borne barley leaf stripe pathogen in the absence of hypersensitive cell death. PLoS ONE. 2010;5:12599. doi: 10.1371/journal.pone.0012599. - DOI - PMC - PubMed
-
- Haegi A., Porta-Puglia A. Purification and partial characterization of a toxic compound produced by Pyrenophora graminea. Physiol. Mol. Plant Pathol. 1995;46:429–444. doi: 10.1006/pmpp.1995.1033. - DOI
-
- Guo M., Si E.J., Hou J.J., Yao L.R., Wang J.C., Meng Y.X., Ma X.L., Li B.C., Wang H.J. Pgmiox mediates stress response and plays a critical role for pathogenicity in Pyrenophora graminea, the agent of barley leaf stripe. Plant Sci. 2025;350:112308. doi: 10.1016/j.plantsci.2024.112308. - DOI - PubMed
Grants and funding
- 32160496/National Natural Science Foundation of China
- 32460517/National Natural Science Foundation of China
- CARS-05-02A-02/China Agriculture Research System of MOF and MARA
- 2021 CYZC-12/Industrial Support Project of Gansu Provincial Education Department
- GSARS-07/Gansu Province's Cold-Dryland Agricultural Triticeae Crops Industry Technology System
- 202501068/Entrepreneurship and Innovation Training Program for College Students of Gansu Agricultural University
- 202501036/Entrepreneurship and Innovation Training Program for College Students of Gansu Agricultural University
- 25CXNA030/The East West Science and Technology Cooperation Special Project
- 25YFNA032/The Key Research and Development Program of Gansu Provincial Department of Science and Technology
- 24JRRA840/The Gansu Province Science and Technology Joint Plan Fund Project
LinkOut - more resources
Full Text Sources

