Construction of Ancestral Chromosomes in Gymnosperms and the Application in Comparative Genomic Analysis
- PMID: 40805710
- PMCID: PMC12349002
- DOI: 10.3390/plants14152361
Construction of Ancestral Chromosomes in Gymnosperms and the Application in Comparative Genomic Analysis
Abstract
Chromosome rearrangements during plant evolution can lead to alterations in genome structure and gene function, thereby influencing species adaptation and evolutionary processes. Gymnosperms, as an ancient group of plants, offer valuable insights into the morphological, physiological, and ecological characteristics of early terrestrial flora. The reconstruction of ancestral karyotypes in gymnosperms may provide critical clues for understanding their evolutionary history. In this study, we inferred the ancestral gymnosperm karyotype (AGK), which comprises 12 chromosomes, and conducted a collinearity analysis with existing gymnosperm genomes. Our findings indicate that chromosome numbers have remained remarkably stable throughout the evolution of gymnosperms. For species with multiplied chromosome numbers, such as gnetophytes, weak collinearities with the AGK were observed. Comparisons between the AGK and gnetophyte genomes revealed a biased pattern regarding retained duplication blocks. Furthermore, our analysis of transposable elements in Welwitschia mirabilis identified enriched regions containing LINE-1 retrotransposons within the syntenic blocks. Syntenic analysis between the AGK and angiosperms also demonstrated a biased distribution across chromosomes. These results provide a fundamental resource for further characterization of chromosomal evolution in gymnosperms.
Keywords: LINE-1; ancestral karyotype; chromosome stability; collinearity; gymnosperm.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
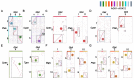
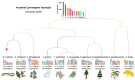

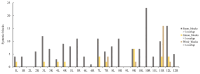

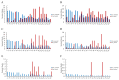
References
-
- Zhang K., Wang X., Cheng F. Plant Polyploidy: Origin, Evolution, and Its Influence on Crop Domestication. Hortic. Plant J. 2019;5:231–239. doi: 10.1016/j.hpj.2019.11.003. - DOI
-
- Liu J., Zhou S.Z., Liu Y.L., Zhao B.Y., Yu D., Zhong M.C., Jiang X.D., Cui W.H., Zhao J.X., Qiu J., et al. Genomes of Meniocus linifolius and Tetracme quadricornis reveal the ancestral karyotype and features of core Brassicaceae. Plant Commun. 2024;5:100878. doi: 10.1016/j.xplc.2024.100878. - DOI - PMC - PubMed
-
- Tkach N., Winterfeld G., Roeser M. Genome sizes of grasses (Poaceae), chromosomal evolution, paleogenomics and the ancestral grass karyotype (AGK) Plant Syst. Evol. 2025;311:4. doi: 10.1007/s00606-024-01934-x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

